
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| HNSC | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ESCA | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| SKCM | ||
| LGG | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000432193 | MMR_HSR1 | PF01926.23 | 2.1e-05 | 1 | 1 |
| ENSP00000437235 | MMR_HSR1 | PF01926.23 | 2.3e-05 | 1 | 1 |
| ENSP00000434953 | MMR_HSR1 | PF01926.23 | 3.3e-05 | 1 | 1 |
| ENSP00000434974 | MMR_HSR1 | PF01926.23 | 4.9e-05 | 1 | 1 |
| ENSP00000260056 | MMR_HSR1 | PF01926.23 | 4.9e-05 | 1 | 1 |
| ENSP00000435089 | MMR_HSR1 | PF01926.23 | 4.9e-05 | 1 | 1 |
| ENSP00000435189 | MMR_HSR1 | PF01926.23 | 4.9e-05 | 1 | 1 |
| ENSP00000478702 | MMR_HSR1 | PF01926.23 | 4.9e-05 | 1 | 1 |
| ENSP00000432336 | MMR_HSR1 | PF01926.23 | 6.4e-05 | 1 | 1 |
| ENSP00000435542 | MMR_HSR1 | PF01926.23 | 0.00011 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000528379 | RAB30-206 | 558 | - | ENSP00000434106 | 55 (aa) | - | E9PRX0 |
| ENST00000532548 | RAB30-209 | 644 | - | ENSP00000437235 | 122 (aa) | - | E9PNB9 |
| ENST00000533276 | RAB30-211 | 548 | - | ENSP00000434528 | 59 (aa) | - | E9PQ07 |
| ENST00000530224 | RAB30-207 | 582 | - | ENSP00000436282 | 37 (aa) | - | E9PPW9 |
| ENST00000534103 | RAB30-213 | 673 | - | ENSP00000435542 | 100 (aa) | - | E9PJQ5 |
| ENST00000527633 | RAB30-205 | 792 | - | ENSP00000435089 | 203 (aa) | - | Q15771 |
| ENST00000526205 | RAB30-204 | 591 | - | ENSP00000432336 | 114 (aa) | - | E9PRF7 |
| ENST00000525117 | RAB30-203 | 937 | - | ENSP00000433243 | 94 (aa) | - | E9PLM3 |
| ENST00000260056 | RAB30-201 | 1393 | - | ENSP00000260056 | 203 (aa) | - | Q15771 |
| ENST00000533486 | RAB30-212 | 9929 | - | ENSP00000435189 | 203 (aa) | - | Q15771 |
| ENST00000533014 | RAB30-210 | 580 | - | ENSP00000433832 | 167 (aa) | - | H0YDK7 |
| ENST00000534141 | RAB30-214 | 3746 | - | ENSP00000434974 | 164 (aa) | - | Q15771 |
| ENST00000612684 | RAB30-216 | 9783 | - | ENSP00000478702 | 203 (aa) | - | Q15771 |
| ENST00000534301 | RAB30-215 | 580 | - | ENSP00000432193 | 139 (aa) | - | E9PS06 |
| ENST00000524635 | RAB30-202 | 495 | - | ENSP00000436587 | 75 (aa) | - | E9PI18 |
| ENST00000531021 | RAB30-208 | 588 | - | ENSP00000434953 | 168 (aa) | - | E9PMJ1 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000137502 | Surveys and Questionnaires | 4E-7 | 22566634 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000137502 | rs11233413 | 11 | 83012030 | T | Economic and political preferences (feminism/equality) | 22566634 | [0.067-0.149] unit decrease | 0.108 | EFO_0004827 |
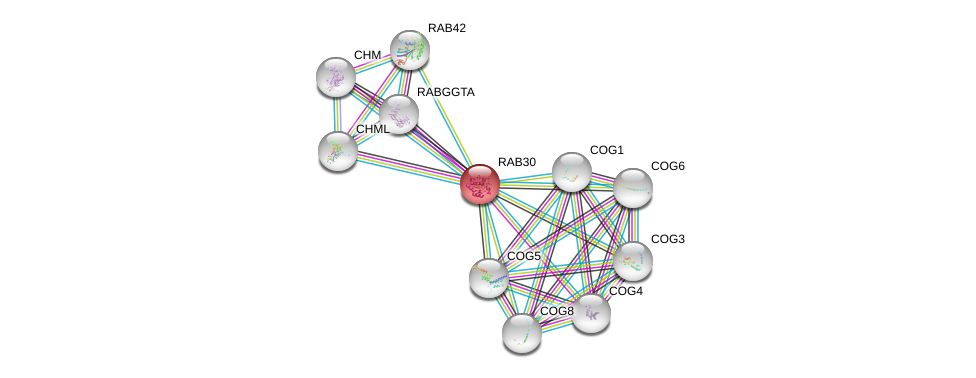
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSAPOG00000022625 | rab30 | 100 | 86.207 | Acanthochromis_polyacanthus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSAMEG00000005231 | RAB30 | 100 | 100.000 | Ailuropoda_melanoleuca |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSAOCG00000012556 | rab30 | 69 | 86.010 | Amphiprion_ocellaris |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSAPEG00000012044 | rab30 | 100 | 86.207 | Amphiprion_percula |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSATEG00000010475 | rab30 | 100 | 86.207 | Anabas_testudineus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSACAG00000001098 | RAB30 | 100 | 98.030 | Anolis_carolinensis |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSANAG00000029292 | RAB30 | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSACLG00000023526 | rab30 | 100 | 86.700 | Astatotilapia_calliptera |
| ENSG00000137502 | RAB30 | 100 | 94.000 | ENSAMXG00000009935 | rab30 | 82 | 85.222 | Astyanax_mexicanus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSBTAG00000002417 | RAB30 | 100 | 100.000 | Bos_taurus |
| ENSG00000137502 | RAB30 | 98 | 77.551 | WBGene00004282 | rab-30 | 81 | 63.218 | Caenorhabditis_elegans |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCJAG00000041773 | RAB30 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCAFG00000004616 | RAB30 | 100 | 100.000 | Canis_familiaris |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCAFG00020017865 | RAB30 | 100 | 100.000 | Canis_lupus_dingo |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCHIG00000020601 | RAB30 | 100 | 100.000 | Capra_hircus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSTSYG00000009392 | RAB30 | 100 | 100.000 | Carlito_syrichta |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCAPG00000009331 | RAB30 | 100 | 100.000 | Cavia_aperea |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCPOG00000005047 | RAB30 | 100 | 100.000 | Cavia_porcellus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCCAG00000026824 | RAB30 | 100 | 100.000 | Cebus_capucinus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCATG00000040441 | RAB30 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCLAG00000014030 | RAB30 | 100 | 100.000 | Chinchilla_lanigera |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCSAG00000003704 | RAB30 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000137502 | RAB30 | 100 | 99.123 | ENSCPBG00000006855 | RAB30 | 100 | 97.537 | Chrysemys_picta_bellii |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCANG00000022240 | RAB30 | 100 | 99.507 | Colobus_angolensis_palliatus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCGRG00001021787 | Rab30 | 100 | 100.000 | Cricetulus_griseus_chok1gshd |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSCGRG00000014842 | Rab30 | 100 | 100.000 | Cricetulus_griseus_crigri |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSCSEG00000007292 | rab30 | 100 | 85.714 | Cynoglossus_semilaevis |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSCVAG00000018085 | rab30 | 100 | 85.222 | Cyprinodon_variegatus |
| ENSG00000137502 | RAB30 | 98 | 95.918 | ENSDARG00000037884 | rab30 | 100 | 85.572 | Danio_rerio |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSDNOG00000039115 | RAB30 | 100 | 100.000 | Dasypus_novemcinctus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSDORG00000010504 | - | 100 | 99.015 | Dipodomys_ordii |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSDORG00000015066 | - | 100 | 97.044 | Dipodomys_ordii |
| ENSG00000137502 | RAB30 | 96 | 92.453 | FBgn0031882 | Rab30 | 87 | 67.692 | Drosophila_melanogaster |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSEASG00005012754 | RAB30 | 100 | 100.000 | Equus_asinus_asinus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSECAG00000022769 | RAB30 | 100 | 100.000 | Equus_caballus |
| ENSG00000137502 | RAB30 | 100 | 95.000 | ENSELUG00000022997 | rab30 | 100 | 85.149 | Esox_lucius |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSFCAG00000014416 | RAB30 | 100 | 100.000 | Felis_catus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSFALG00000007765 | RAB30 | 100 | 98.522 | Ficedula_albicollis |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSFDAG00000014521 | RAB30 | 100 | 100.000 | Fukomys_damarensis |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSFHEG00000013918 | rab30 | 100 | 85.714 | Fundulus_heteroclitus |
| ENSG00000137502 | RAB30 | 100 | 96.364 | ENSGMOG00000002552 | rab30 | 100 | 81.773 | Gadus_morhua |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSGALG00000017248 | RAB30 | 100 | 98.522 | Gallus_gallus |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSGAFG00000020309 | rab30 | 100 | 85.222 | Gambusia_affinis |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSGACG00000012006 | rab30 | 100 | 87.192 | Gasterosteus_aculeatus |
| ENSG00000137502 | RAB30 | 100 | 99.123 | ENSGAGG00000004474 | RAB30 | 100 | 97.537 | Gopherus_agassizii |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSGGOG00000017157 | RAB30 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSHBUG00000000993 | rab30 | 93 | 86.700 | Haplochromis_burtoni |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSHGLG00000002759 | RAB30 | 100 | 100.000 | Heterocephalus_glaber_female |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSHGLG00100001213 | RAB30 | 100 | 100.000 | Heterocephalus_glaber_male |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSHCOG00000010707 | rab30 | 71 | 89.474 | Hippocampus_comes |
| ENSG00000137502 | RAB30 | 98 | 95.918 | ENSIPUG00000021010 | Rab30 | 100 | 87.562 | Ictalurus_punctatus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSSTOG00000020966 | RAB30 | 100 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSJJAG00000020064 | Rab30 | 100 | 100.000 | Jaculus_jaculus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSKMAG00000004439 | rab30 | 100 | 85.714 | Kryptolebias_marmoratus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSLBEG00000001146 | rab30 | 100 | 86.207 | Labrus_bergylta |
| ENSG00000137502 | RAB30 | 100 | 98.000 | ENSLACG00000006407 | RAB30 | 100 | 91.626 | Latimeria_chalumnae |
| ENSG00000137502 | RAB30 | 100 | 98.000 | ENSLOCG00000005884 | rab30 | 100 | 89.163 | Lepisosteus_oculatus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSLAFG00000010711 | RAB30 | 100 | 99.507 | Loxodonta_africana |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMFAG00000033119 | RAB30 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMMUG00000007842 | RAB30 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMNEG00000037406 | RAB30 | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMLEG00000039496 | RAB30 | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000137502 | RAB30 | 100 | 96.000 | ENSMAMG00000011973 | rab30 | 100 | 86.207 | Mastacembelus_armatus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSMZEG00005024032 | rab30 | 93 | 86.700 | Maylandia_zebra |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMGAG00000015230 | RAB30 | 100 | 97.549 | Meleagris_gallopavo |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMAUG00000008179 | Rab30 | 100 | 100.000 | Mesocricetus_auratus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMICG00000012002 | RAB30 | 100 | 100.000 | Microcebus_murinus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMOCG00000009821 | - | 100 | 100.000 | Microtus_ochrogaster |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSMMOG00000000304 | rab30 | 100 | 85.714 | Mola_mola |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMODG00000004506 | RAB30 | 100 | 99.507 | Monodelphis_domestica |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSMALG00000010438 | rab30 | 100 | 86.700 | Monopterus_albus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | MGP_CAROLIEiJ_G0029992 | Rab30 | 100 | 99.507 | Mus_caroli |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMUSG00000030643 | Rab30 | 100 | 100.000 | Mus_musculus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | MGP_PahariEiJ_G0013212 | Rab30 | 100 | 100.000 | Mus_pahari |
| ENSG00000137502 | RAB30 | 100 | 100.000 | MGP_SPRETEiJ_G0031102 | Rab30 | 100 | 100.000 | Mus_spretus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMPUG00000008088 | RAB30 | 100 | 100.000 | Mustela_putorius_furo |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMLUG00000006552 | RAB30 | 100 | 98.522 | Myotis_lucifugus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSNGAG00000018930 | Rab30 | 100 | 100.000 | Nannospalax_galili |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSNBRG00000002082 | rab30 | 93 | 86.700 | Neolamprologus_brichardi |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSNLEG00000029364 | RAB30 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSMEUG00000012886 | RAB30 | 100 | 97.537 | Notamacropus_eugenii |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSOPRG00000007511 | RAB30 | 100 | 100.000 | Ochotona_princeps |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSODEG00000003833 | RAB30 | 100 | 100.000 | Octodon_degus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSONIG00000005708 | rab30 | 100 | 86.700 | Oreochromis_niloticus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSOCUG00000007580 | RAB30 | 100 | 100.000 | Oryctolagus_cuniculus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSORLG00000029812 | rab30 | 100 | 85.714 | Oryzias_latipes |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSORLG00020015288 | rab30 | 100 | 85.714 | Oryzias_latipes_hni |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSORLG00015020972 | rab30 | 100 | 85.714 | Oryzias_latipes_hsok |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSOMEG00000001613 | rab30 | 100 | 85.222 | Oryzias_melastigma |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSOGAG00000033189 | RAB30 | 100 | 100.000 | Otolemur_garnettii |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSOARG00000005688 | RAB30 | 100 | 100.000 | Ovis_aries |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPPAG00000031604 | RAB30 | 100 | 100.000 | Pan_paniscus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPPRG00000023881 | RAB30 | 100 | 100.000 | Panthera_pardus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPTIG00000009836 | RAB30 | 100 | 100.000 | Panthera_tigris_altaica |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPTRG00000004125 | RAB30 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPANG00000003974 | RAB30 | 100 | 100.000 | Papio_anubis |
| ENSG00000137502 | RAB30 | 100 | 96.364 | ENSPKIG00000015202 | rab30 | 99 | 88.614 | Paramormyrops_kingsleyae |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPSIG00000003078 | RAB30 | 100 | 96.552 | Pelodiscus_sinensis |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSPMGG00000016093 | rab30 | 100 | 76.293 | Periophthalmus_magnuspinnatus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPEMG00000023854 | Rab30 | 100 | 100.000 | Peromyscus_maniculatus_bairdii |
| ENSG00000137502 | RAB30 | 96 | 92.453 | ENSPMAG00000006105 | rab30 | 99 | 67.619 | Petromyzon_marinus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPCIG00000027547 | RAB30 | 94 | 99.507 | Phascolarctos_cinereus |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSPFOG00000005479 | rab30 | 100 | 85.714 | Poecilia_formosa |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSPLAG00000015383 | rab30 | 100 | 85.714 | Poecilia_latipinna |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSPMEG00000009701 | rab30 | 100 | 85.714 | Poecilia_mexicana |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSPREG00000014634 | rab30 | 100 | 85.714 | Poecilia_reticulata |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPPYG00000003733 | RAB30 | 100 | 100.000 | Pongo_abelii |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPCAG00000000574 | RAB30 | 100 | 99.507 | Procavia_capensis |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPCOG00000017316 | RAB30 | 100 | 100.000 | Propithecus_coquereli |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSPVAG00000005455 | RAB30 | 100 | 100.000 | Pteropus_vampyrus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSPNYG00000009728 | rab30 | 93 | 86.207 | Pundamilia_nyererei |
| ENSG00000137502 | RAB30 | 98 | 95.918 | ENSPNAG00000010760 | rab30 | 100 | 87.065 | Pygocentrus_nattereri |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSRNOG00000010224 | Rab30 | 100 | 100.000 | Rattus_norvegicus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSRBIG00000039227 | RAB30 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSRROG00000031260 | RAB30 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSSBOG00000026403 | RAB30 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSSHAG00000018081 | RAB30 | 100 | 98.058 | Sarcophilus_harrisii |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSSMAG00000002500 | rab30 | 100 | 85.714 | Scophthalmus_maximus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSSDUG00000013325 | rab30 | 100 | 86.700 | Seriola_dumerili |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSSLDG00000022533 | rab30 | 100 | 86.700 | Seriola_lalandi_dorsalis |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSSPUG00000006349 | RAB30 | 100 | 97.044 | Sphenodon_punctatus |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSSPAG00000013422 | rab30 | 100 | 86.700 | Stegastes_partitus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSSSCG00000014900 | RAB30 | 100 | 100.000 | Sus_scrofa |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSTGUG00000012941 | RAB30 | 100 | 98.522 | Taeniopygia_guttata |
| ENSG00000137502 | RAB30 | 100 | 97.000 | ENSTRUG00000022559 | rab30 | 99 | 86.070 | Takifugu_rubripes |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSTBEG00000009832 | RAB30 | 100 | 100.000 | Tupaia_belangeri |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSTTRG00000006100 | RAB30 | 100 | 100.000 | Tursiops_truncatus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSUAMG00000009326 | RAB30 | 100 | 100.000 | Ursus_americanus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSUMAG00000023572 | RAB30 | 100 | 100.000 | Ursus_maritimus |
| ENSG00000137502 | RAB30 | 100 | 100.000 | ENSVVUG00000017471 | RAB30 | 100 | 100.000 | Vulpes_vulpes |
| ENSG00000137502 | RAB30 | 100 | 98.182 | ENSXETG00000013633 | rab30 | 100 | 93.103 | Xenopus_tropicalis |
| ENSG00000137502 | RAB30 | 100 | 98.305 | ENSXMAG00000025848 | rab30 | 100 | 85.714 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000139 | Golgi membrane | 21873635. | IBA | Component |
| GO:0000139 | Golgi membrane | - | TAS | Component |
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005789 | endoplasmic reticulum membrane | 21873635. | IBA | Component |
| GO:0005795 | Golgi stack | 9792283. | TAS | Component |
| GO:0005801 | cis-Golgi network | 22188167. | IDA | Component |
| GO:0005802 | trans-Golgi network | 21873635. | IBA | Component |
| GO:0005802 | trans-Golgi network | 22188167. | IDA | Component |
| GO:0006886 | intracellular protein transport | 21873635. | IBA | Process |
| GO:0006888 | ER to Golgi vesicle-mediated transport | 21873635. | IBA | Process |
| GO:0007030 | Golgi organization | 22188167. | IMP | Process |
| GO:0031985 | Golgi cisterna | 22188167. | IDA | Component |
| GO:0032482 | Rab protein signal transduction | 21873635. | IBA | Process |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |