
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| BRCA | |||
| CESC | |||
| CHOL | |||
| ESCA | |||
| HNSC | |||
| KIRP | |||
| LIHC | |||
| LUAD | |||
| MESO | |||
| PRAD | |||
| READ | |||
| SARC | |||
| STAD | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| ACC | ||
| KIRP | ||
| READ | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| LUAD |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000341980 | PPP2R1B-202 | 1953 | - | ENSP00000343317 | 556 (aa) | - | P30154 |
| ENST00000426998 | PPP2R1B-204 | 1888 | - | ENSP00000410671 | 603 (aa) | - | P30154 |
| ENST00000393055 | PPP2R1B-203 | 1525 | - | ENSP00000376775 | 474 (aa) | - | P30154 |
| ENST00000527614 | PPP2R1B-206 | 5587 | XM_017017961 | ENSP00000437193 | 601 (aa) | XP_016873450 | P30154 |
| ENST00000531890 | PPP2R1B-210 | 670 | - | ENSP00000433598 | 224 (aa) | - | H0YDG7 |
| ENST00000531373 | PPP2R1B-209 | 553 | - | ENSP00000434705 | 119 (aa) | - | E9PNM7 |
| ENST00000534500 | PPP2R1B-211 | 582 | - | ENSP00000432666 | 83 (aa) | - | E9PPI5 |
| ENST00000530787 | PPP2R1B-208 | 131 | - | - | - (aa) | - | - |
| ENST00000529672 | PPP2R1B-207 | 874 | - | - | - (aa) | - | - |
| ENST00000534521 | PPP2R1B-212 | 2294 | - | ENSP00000436626 | 135 (aa) | - | E9PHZ6 |
| ENST00000311129 | PPP2R1B-201 | 2082 | XM_017017960 | ENSP00000311344 | 667 (aa) | XP_016873449 | P30154 |
| ENST00000526287 | PPP2R1B-205 | 101 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03015 | mRNA surveillance pathway | KEGG |
| hsa04071 | Sphingolipid signaling pathway | KEGG |
| hsa04114 | Oocyte meiosis | KEGG |
| hsa04151 | PI3K-Akt signaling pathway | KEGG |
| hsa04152 | AMPK signaling pathway | KEGG |
| hsa04261 | Adrenergic signaling in cardiomyocytes | KEGG |
| hsa04350 | TGF-beta signaling pathway | KEGG |
| hsa04390 | Hippo signaling pathway | KEGG |
| hsa04530 | Tight junction | KEGG |
| hsa04728 | Dopaminergic synapse | KEGG |
| hsa04730 | Long-term depression | KEGG |
| hsa05142 | Chagas disease (American trypanosomiasis) | KEGG |
| hsa05160 | Hepatitis C | KEGG |
| hsa05165 | Human papillomavirus infection | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000137713 | Myocardial Infarction | 2.1990000E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000137713 | rs11213999 | 11 | 111763868 | A | Facial wrinkles | 29555444 | [NR] z score decrease | 5.04 | HP_0009762 |
| ENSG00000137713 | rs4936675 | 11 | 111752364 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 | ||
| ENSG00000137713 | rs643506 | 11 | 111765903 | G | Breast cancer | 29059683 | [0.021-0.047] unit decrease | 0.0344 | EFO_0000305 |
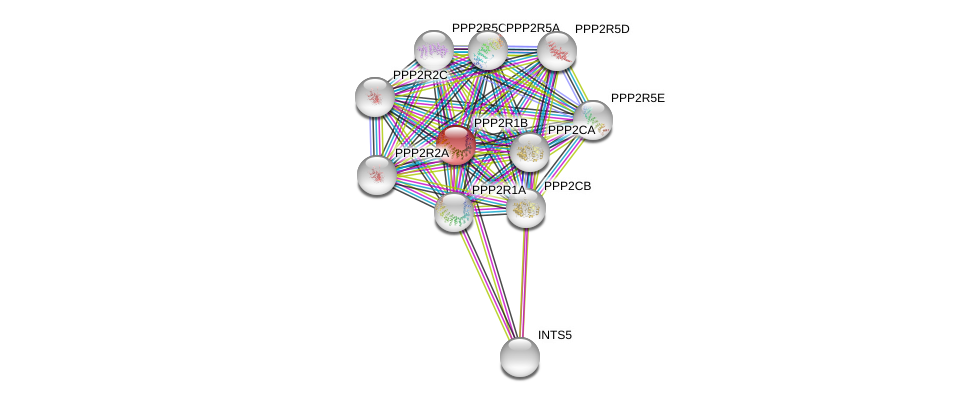
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000137713 | PPP2R1B | 79 | 85.106 | WBGene00003901 | paa-1 | 98 | 62.544 | Caenorhabditis_elegans |
| ENSG00000137713 | PPP2R1B | 96 | 71.848 | FBgn0260439 | Pp2A-29B | 98 | 72.021 | Drosophila_melanogaster |
| ENSG00000137713 | PPP2R1B | 98 | 86.224 | ENSG00000105568 | PPP2R1A | 99 | 86.403 | Homo_sapiens |
| ENSG00000137713 | PPP2R1B | 97 | 96.207 | ENSMUSG00000032058 | Ppp2r1b | 100 | 95.674 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000159 | protein phosphatase type 2A complex | 21873635. | IBA | Component |
| GO:0004722 | protein serine/threonine phosphatase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 8392071.16456541.17540176.18715871.18782753.19156129.21172653.23555304.26496610.28330616. | IPI | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0006470 | protein dephosphorylation | - | IEA | Process |
| GO:0019888 | protein phosphatase regulator activity | 21873635. | IBA | Function |
| GO:0043666 | regulation of phosphoprotein phosphatase activity | - | IEA | Process |
| GO:0045121 | membrane raft | 21172653. | IDA | Component |
| GO:0060561 | apoptotic process involved in morphogenesis | 21172653. | IMP | Process |
| GO:0070062 | extracellular exosome | 23533145. | HDA | Component |
| GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 21172653. | IMP | Process |