
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| COAD | |||
| MESO | |||
| PAAD | |||
| READ | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| LUSC | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| CESC | ||
| KICH | ||
| LIHC | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000445435 | CEP55-202 | 747 | - | ENSP00000389150 | 208 (aa) | - | H0Y432 |
| ENST00000496302 | CEP55-203 | 710 | - | - | - (aa) | - | - |
| ENST00000371485 | CEP55-201 | 2638 | XM_011539918 | ENSP00000360540 | 464 (aa) | XP_011538220 | Q53EZ4 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000138180 | Glomerular Filtration Rate | 1.2719533E-005 | - |
| ENSG00000138180 | Glomerular Filtration Rate | 8.7220442E-005 | - |
| ENSG00000138180 | Glomerular Filtration Rate | 8.1242576E-005 | - |
| ENSG00000138180 | Platelet Function Tests | 4.5925000E-006 | - |
| ENSG00000138180 | Platelet Function Tests | 1.4019000E-006 | - |
| ENSG00000138180 | Platelet Function Tests | 1.5923000E-005 | - |
| ENSG00000138180 | Platelet Function Tests | 1.5923000E-005 | - |
| ENSG00000138180 | Platelet Function Tests | 1.4892000E-005 | - |
| ENSG00000138180 | Bronchodilator Agents | 3E-6 | 26634245 |
| ENSG00000138180 | Lung Volume Measurements | 3E-6 | 26634245 |
| ENSG00000138180 | Bronchodilator Agents | 3E-6 | 26634245 |
| ENSG00000138180 | Lung Volume Measurements | 3E-6 | 26634245 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000138180 | rs10882258 | 10 | 93515133 | ? | Lobe attachment (rater-scored or self-reported) | 29198719 | EFO_0007667 |
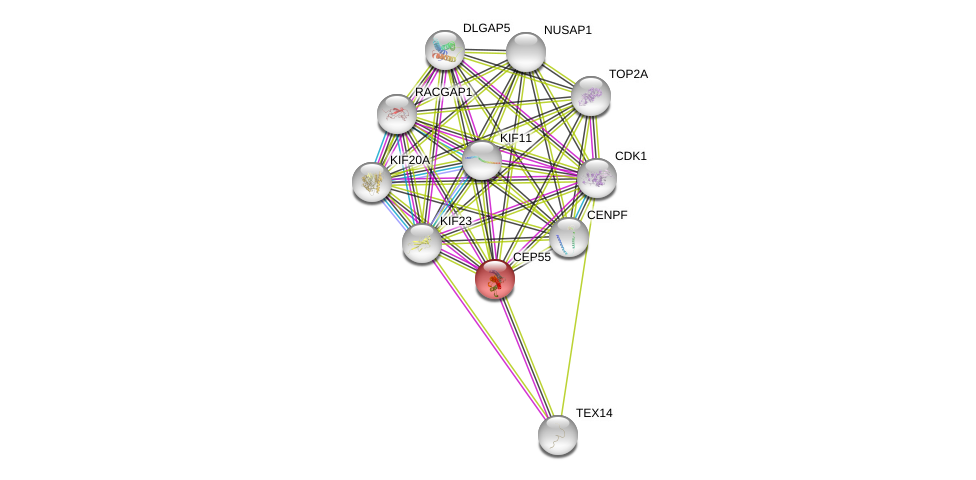
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000281 | mitotic cytokinesis | 21873635. | IBA | Process |
| GO:0000281 | mitotic cytokinesis | 19638580. | IGI | Process |
| GO:0005515 | protein binding | 16189514.17853893.18940611.19549727.20176808.21516116.25416956.25910212.26638075.26871637.27107012. | IPI | Function |
| GO:0005813 | centrosome | 20186884.21399614. | IDA | Component |
| GO:0005814 | centriole | - | IEA | Component |
| GO:0005815 | microtubule organizing center | - | IDA | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0006997 | nucleus organization | 20616062. | IMP | Process |
| GO:0007080 | mitotic metaphase plate congression | 20616062. | IMP | Process |
| GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0030496 | midbody | 21873635. | IBA | Component |
| GO:0030496 | midbody | 20186884.21310966.28264986. | IDA | Component |
| GO:0032154 | cleavage furrow | - | IEA | Component |
| GO:0045171 | intercellular bridge | - | IEA | Component |
| GO:0045184 | establishment of protein localization | 21873635. | IBA | Process |
| GO:0045184 | establishment of protein localization | 23045692. | IMP | Process |
| GO:0061952 | midbody abscission | 20616062. | IMP | Process |
| GO:0072001 | renal system development | 28264986. | IMP | Process |
| GO:0090543 | Flemming body | 18641129. | IDA | Component |
| GO:1904888 | cranial skeletal system development | 28264986. | IMP | Process |