
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| READ | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| UCS | ||
| SKCM | ||
| KIRP | ||
| BLCA | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000372921 | ANXA7-202 | 2452 | XM_017016163 | ENSP00000362012 | 466 (aa) | XP_016871652 | P20073 |
| ENST00000463788 | ANXA7-204 | 739 | - | - | - (aa) | - | - |
| ENST00000372919 | ANXA7-201 | 2174 | XM_017016162 | ENSP00000362010 | 488 (aa) | XP_016871651 | P20073 |
| ENST00000394847 | ANXA7-203 | 648 | - | ENSP00000378317 | 144 (aa) | - | B9ZVT2 |
| ENST00000492380 | ANXA7-205 | 843 | - | - | - (aa) | - | - |
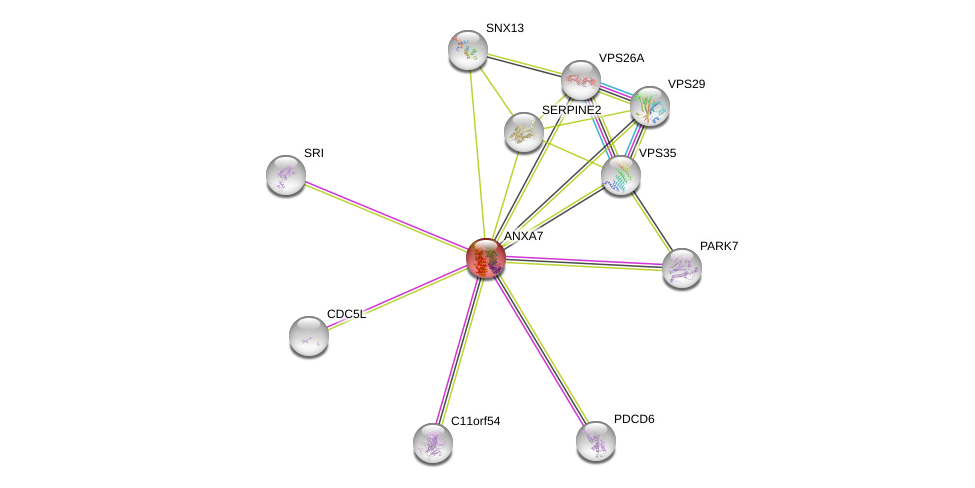
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000138279 | ANXA7 | 96 | 81.210 | ENSMUSG00000021814 | Anxa7 | 100 | 89.535 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0005178 | integrin binding | 21873635. | IBA | Function |
| GO:0005178 | integrin binding | 24007983. | IPI | Function |
| GO:0005509 | calcium ion binding | - | IEA | Function |
| GO:0005515 | protein binding | 9268363.12445460.17699613.18256029. | IPI | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 21873635. | IBA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 10672515. | IDA | Component |
| GO:0005635 | nuclear envelope | - | IEA | Component |
| GO:0005789 | endoplasmic reticulum membrane | 10672515. | IDA | Component |
| GO:0005829 | cytosol | - | IEA | Component |
| GO:0005886 | plasma membrane | - | IEA | Component |
| GO:0006874 | cellular calcium ion homeostasis | 21873635. | IBA | Process |
| GO:0006914 | autophagy | 24007983. | IMP | Process |
| GO:0007599 | hemostasis | - | IEA | Process |
| GO:0008283 | cell proliferation | - | IEA | Process |
| GO:0008360 | regulation of cell shape | - | IEA | Process |
| GO:0009651 | response to salt stress | - | IEA | Process |
| GO:0009992 | cellular water homeostasis | - | IEA | Process |
| GO:0010629 | negative regulation of gene expression | 24007983. | IMP | Process |
| GO:0014070 | response to organic cyclic compound | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0030855 | epithelial cell differentiation | 21873635. | IBA | Process |
| GO:0030855 | epithelial cell differentiation | 21492153. | IEP | Process |
| GO:0035176 | social behavior | 21531385. | IEP | Process |
| GO:0042584 | chromaffin granule membrane | 21873635. | IBA | Component |
| GO:0042584 | chromaffin granule membrane | 9268363. | IDA | Component |
| GO:0042802 | identical protein binding | 21873635. | IBA | Function |
| GO:0048306 | calcium-dependent protein binding | 21873635. | IBA | Function |
| GO:0048306 | calcium-dependent protein binding | 12445460. | IPI | Function |
| GO:0051592 | response to calcium ion | 21873635. | IBA | Process |
| GO:0061025 | membrane fusion | - | IEA | Process |
| GO:0062023 | collagen-containing extracellular matrix | 25037231. | HDA | Component |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.23533145. | HDA | Component |