
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| MESO | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| KIRP | |||
| LGG | |||
| SARC | |||
| SKCM | |||
| THCA | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| ACC | ||
| SKCM | ||
| LUSC | ||
| PAAD | ||
| BLCA | ||
| LAML | ||
| KICH | ||
| LIHC | ||
| LUAD |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000544334 | RPTOR-202 | 5734 | - | ENSP00000442479 | 1177 (aa) | - | Q8N122 |
| ENST00000306801 | RPTOR-201 | 6408 | - | ENSP00000307272 | 1335 (aa) | - | Q8N122 |
| ENST00000575542 | RPTOR-209 | 5536 | - | - | - (aa) | - | - |
| ENST00000574767 | RPTOR-208 | 2461 | - | ENSP00000459701 | 95 (aa) | - | I3L2I3 |
| ENST00000649732 | RPTOR-212 | 2499 | - | - | - (aa) | - | - |
| ENST00000577161 | RPTOR-211 | 3633 | - | - | - (aa) | - | - |
| ENST00000572733 | RPTOR-204 | 424 | - | - | - (aa) | - | - |
| ENST00000573746 | RPTOR-206 | 688 | - | - | - (aa) | - | - |
| ENST00000570891 | RPTOR-203 | 2286 | - | ENSP00000460136 | 379 (aa) | - | Q8N122 |
| ENST00000573163 | RPTOR-205 | 501 | - | - | - (aa) | - | - |
| ENST00000573837 | RPTOR-207 | 443 | - | - | - (aa) | - | - |
| ENST00000576366 | RPTOR-210 | 787 | - | ENSP00000460940 | 235 (aa) | - | I3L436 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04136 | Autophagy - other | KEGG |
| hsa04140 | Autophagy - animal | KEGG |
| hsa04150 | mTOR signaling pathway | KEGG |
| hsa04151 | PI3K-Akt signaling pathway | KEGG |
| hsa04152 | AMPK signaling pathway | KEGG |
| hsa04211 | Longevity regulating pathway | KEGG |
| hsa04213 | Longevity regulating pathway - multiple species | KEGG |
| hsa04714 | Thermogenesis | KEGG |
| hsa04910 | Insulin signaling pathway | KEGG |
| hsa05206 | MicroRNAs in cancer | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000141564 | Blood Pressure | 3.0510000E-005 | - |
| ENSG00000141564 | Blood Pressure | 9.4100000E-005 | - |
| ENSG00000141564 | Blood Pressure | 1.2750000E-005 | - |
| ENSG00000141564 | Blood Pressure | 7.9020000E-005 | - |
| ENSG00000141564 | Blood Pressure | 8.9430000E-005 | - |
| ENSG00000141564 | Blood Pressure | 6.9960000E-005 | - |
| ENSG00000141564 | Platelet Function Tests | 2.2304700E-005 | - |
| ENSG00000141564 | Platelet Function Tests | 1.3048300E-005 | - |
| ENSG00000141564 | Platelet Function Tests | 2.8965900E-005 | - |
| ENSG00000141564 | Platelet Function Tests | 3.1423611E-006 | - |
| ENSG00000141564 | Platelet Function Tests | 4.1482005E-006 | - |
| ENSG00000141564 | Platelet Function Tests | 2.6898337E-006 | - |
| ENSG00000141564 | Platelet Function Tests | 1.6369963E-006 | - |
| ENSG00000141564 | Platelet Function Tests | 7.5636642E-007 | - |
| ENSG00000141564 | Schizophrenia | 7E-6 | 26198764 |
| ENSG00000141564 | Bronchial Provocation Tests | 9E-6 | 25514360 |
| ENSG00000141564 | Pulmonary Disease, Chronic Obstructive | 9E-6 | 25514360 |
| ENSG00000141564 | Body Mass Index | 5E-7 | 25673413 |
| ENSG00000141564 | Body Mass Index | 2E-9 | 25673413 |
| ENSG00000141564 | Myopia, Degenerative | 2E-6 | 23049088 |
| ENSG00000141564 | Body Mass Index | 4E-10 | 25673413 |
| ENSG00000141564 | Obesity | 2E-8 | 23563607 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000141564 | rs4969427 | 17 | 80789086 | A | Cognitive empathy | 28584286 | [0.09-0.222] unit decrease | 0.1559 | EFO_0009183 |
| ENSG00000141564 | rs139293840 | 17 | 80556559 | ? | Pulse pressure | 29403010 | [0.074-0.141] unit increase | 0.1079 | EFO_0005763 |
| ENSG00000141564 | rs8066384 | 17 | 80651956 | C | Schizophrenia | 26198764 | [NR] | 1.0526316 | EFO_0000692 |
| ENSG00000141564 | rs7215564 | 17 | 80763487 | ? | Myopia (pathological) | 23049088 | [NR] | EFO_0004207 | |
| ENSG00000141564 | rs7503807 | 17 | 80617311 | A | Obesity | 23563607 | [NR] | 1.04 | EFO_0001073 |
| ENSG00000141564 | rs12940622 | 17 | 80641771 | A | Body mass index | 28892062 | [0.01-0.018] kg/m2 decrease | 0.014 | EFO_0004340 |
| ENSG00000141564 | rs4969426 | 17 | 80789008 | T | Moyamoya disease | 29273593 | [NR] | 1.43 | Orphanet_2573 |
| ENSG00000141564 | rs1045626 | 17 | 80966164 | T | Educational attainment (MTAG) | 30038396 | [0.0051-0.0101] unit increase | 0.0076 | EFO_0004784 |
| ENSG00000141564 | rs62069681 | 17 | 80650902 | C | Blood protein levels | 29875488 | [0.43-0.59] unit increase | 0.51 | EFO_0007937 |
| ENSG00000141564 | rs2672886 | 17 | 80807851 | C | Airway responsiveness in chronic obstructive pulmonary disease | 25514360 | [NR] unit increase | 0.051 | EFO_0006897|EFO_0000341 |
| ENSG00000141564 | rs12940622 | 17 | 80641771 | A | BMI (adjusted for smoking behaviour) | 28443625 | [0.014-0.032] kg/m2 decrease | 0.0232 | EFO_0004318|EFO_0004340 |
| ENSG00000141564 | rs12940622 | 17 | 80641771 | A | BMI (adjusted for smoking behaviour) | 28443625 | [0.012-0.024] kg/m2 decrease | 0.0178 | EFO_0004318|EFO_0004340 |
| ENSG00000141564 | rs12940622 | 17 | 80641771 | A | Body mass index (joint analysis main effects and smoking interaction) | 28443625 | EFO_0004318|EFO_0004340 | ||
| ENSG00000141564 | rs12940622 | 17 | 80641771 | A | Body mass index (joint analysis main effects and smoking interaction) | 28443625 | EFO_0004318|EFO_0004340 | ||
| ENSG00000141564 | rs12940622 | 17 | 80641771 | A | BMI in non-smokers | 28443625 | [0.015-0.036] kg/m2 decrease | 0.0258 | EFO_0004340 |
| ENSG00000141564 | rs12940622 | 17 | 80641771 | A | BMI in non-smokers | 28443625 | [0.011-0.025] kg/m2 decrease | 0.0182 | EFO_0004340 |
| ENSG00000141564 | rs12939549 | 17 | 80637924 | A | Body mass index | 26426971 | unit increase | 0.017547995 | EFO_0004340 |
| ENSG00000141564 | rs12939549 | 17 | 80637924 | A | Body mass index | 30108127 | [NR] unit increase | 0.017 | EFO_0004340 |
| ENSG00000141564 | rs58418838 | 17 | 80727155 | ? | Menarche (age at onset) | 30595370 | EFO_0004703 | ||
| ENSG00000141564 | rs11150745 | 17 | 80783826 | ? | Body mass index | 30595370 | EFO_0004340 | ||
| ENSG00000141564 | rs10438820 | 17 | 80550797 | T | Alcohol consumption (drinks per week) (MTAG) | 30643251 | [0.0057-0.0113] unit increase | 0.00850176 | EFO_0007878 |
| ENSG00000141564 | rs10438820 | 17 | 80550797 | T | Alcohol consumption (drinks per week) | 30643251 | [0.0058-0.0121] unit increase | 0.008971778 | EFO_0007878 |
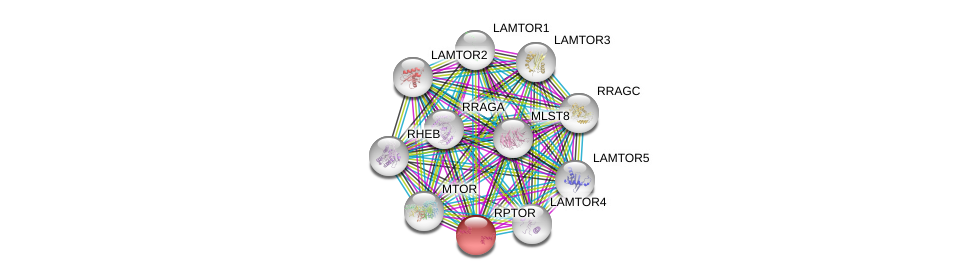
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001030 | RNA polymerase III type 1 promoter DNA binding | 20233713. | IDA | Function |
| GO:0001031 | RNA polymerase III type 2 promoter DNA binding | 20233713. | IDA | Function |
| GO:0001032 | RNA polymerase III type 3 promoter DNA binding | 20233713. | IDA | Function |
| GO:0001156 | TFIIIC-class transcription factor complex binding | 20543138. | IDA | Function |
| GO:0001558 | regulation of cell growth | 20381137. | IMP | Process |
| GO:0001938 | positive regulation of endothelial cell proliferation | - | IEA | Process |
| GO:0005515 | protein binding | 12150925.12150926.12408816.17386266.17461779.17565979.18337751.18955708.19446321.19619545.19875983.20347422.20543138.20562859.21045808.21339740.21471201.21614075.22424946.23027611.23263282.23953116.24036451.24375676.25446900.25561175.25567906.25657994.25686248.25940091.26348909.26678875.26787466.27043084. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 20381137. | IDA | Component |
| GO:0005764 | lysosome | 20381137. | IDA | Component |
| GO:0005764 | lysosome | 22424946. | IMP | Component |
| GO:0005765 | lysosomal membrane | 17897319. | HDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0007050 | cell cycle arrest | - | TAS | Process |
| GO:0008361 | regulation of cell size | 21873635. | IBA | Process |
| GO:0008361 | regulation of cell size | 12150925. | IMP | Process |
| GO:0009267 | cellular response to starvation | 21873635. | IBA | Process |
| GO:0010494 | cytoplasmic stress granule | 25940091. | IDA | Component |
| GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 18426977. | IMP | Process |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016241 | regulation of macroautophagy | - | TAS | Process |
| GO:0019901 | protein kinase binding | 18439900. | IPI | Function |
| GO:0030291 | protein serine/threonine kinase inhibitor activity | 12718876. | IDA | Function |
| GO:0030295 | protein kinase activator activity | 12150926. | IDA | Function |
| GO:0030307 | positive regulation of cell growth | 21873635. | IBA | Process |
| GO:0030307 | positive regulation of cell growth | 18426977. | IMP | Process |
| GO:0030425 | dendrite | - | IEA | Component |
| GO:0030674 | protein binding, bridging | 21873635. | IBA | Function |
| GO:0030674 | protein binding, bridging | 12150926. | IDA | Function |
| GO:0031669 | cellular response to nutrient levels | 12150925. | IMP | Process |
| GO:0031929 | TOR signaling | 21873635. | IBA | Process |
| GO:0031929 | TOR signaling | 20381137. | IDA | Process |
| GO:0031931 | TORC1 complex | 21873635. | IBA | Component |
| GO:0031931 | TORC1 complex | 12150926.12718876.15467718.24036451.25940091. | IDA | Component |
| GO:0032008 | positive regulation of TOR signaling | 12150926.18439900. | IDA | Process |
| GO:0032147 | activation of protein kinase activity | - | IEA | Process |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 18426977. | IMP | Process |
| GO:0038202 | TORC1 signaling | 12718876. | IMP | Process |
| GO:0043025 | neuronal cell body | - | IEA | Component |
| GO:0044877 | protein-containing complex binding | 19211835. | IPI | Function |
| GO:0045945 | positive regulation of transcription by RNA polymerase III | 20233713. | IMP | Process |
| GO:0071230 | cellular response to amino acid stimulus | 21873635. | IBA | Process |
| GO:0071230 | cellular response to amino acid stimulus | 20381137. | IMP | Process |
| GO:0071233 | cellular response to leucine | 22424946. | IDA | Process |
| GO:0071889 | 14-3-3 protein binding | 18439900. | IDA | Function |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | - | IEA | Process |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | 21873635. | IBA | Process |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | 12150926. | IDA | Process |
| GO:1900034 | regulation of cellular response to heat | - | TAS | Process |
| GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 18426977. | IMP | Process |