
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| KIRP | ||
| PCPG | ||
| UCEC | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000411860 | RIO1 | PF01163.22 | 2e-05 | 1 | 1 |
| ENSP00000270112 | RIO1 | PF01163.22 | 0.00013 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000439107 | HUNK-203 | 860 | - | ENSP00000408219 | 112 (aa) | - | H7C2X2 |
| ENST00000270112 | HUNK-201 | 7385 | XM_011529537 | ENSP00000270112 | 714 (aa) | XP_011527839 | P57058 |
| ENST00000430354 | HUNK-202 | 486 | - | ENSP00000411860 | 162 (aa) | - | H7C3H0 |
| ENST00000465574 | HUNK-204 | 832 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000142149 | Hemoglobin A, Glycosylated | 7.84783516711851E-5 | 17903298 |
| ENSG00000142149 | Neuroblastoma | 7.73E-7 | 18463370 |
| ENSG00000142149 | Blood Pressure | 5.6210000E-005 | - |
| ENSG00000142149 | Platelet Function Tests | 2.0700000E-006 | - |
| ENSG00000142149 | Child Development Disorders, Pervasive | 2.0214700E-005 | - |
| ENSG00000142149 | Child Development Disorders, Pervasive | 4.3401000E-005 | - |
| ENSG00000142149 | Child Development Disorders, Pervasive | 2.7675200E-005 | - |
| ENSG00000142149 | Child Development Disorders, Pervasive | 6.0977400E-005 | - |
| ENSG00000142149 | Child Development Disorders, Pervasive | 9.4000000E-005 | - |
| ENSG00000142149 | Interleukin-8 | 1.5900000E-005 | - |
| ENSG00000142149 | Immunoglobulins | 8E-6 | 23382691 |
| ENSG00000142149 | Vitiligo | 2E-6 | 19890347 |
| ENSG00000142149 | Logic | 5E-7 | 21107309 |
| ENSG00000142149 | Diabetes Mellitus, Type 2 | 4E-6 | 21490949 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000142149 | rs11910494 | 21 | 31923801 | G | IgG glycosylation | 23382691 | [0.083-0.213] unit increase | 0.1483 | EFO_0005193 |
| ENSG00000142149 | rs2833607 | 21 | 32008727 | ? | Vitiligo | 19890347 | EFO_0004208 | ||
| ENSG00000142149 | rs2833560 | 21 | 31915561 | ? | Measles | 28928442 | [0.059-0.142] unit increase | 0.1009 | EFO_0008414 |
| ENSG00000142149 | rs2833556 | 21 | 31912988 | ? | Response to antipsychotic treatment in schizophrenia (reasoning) | 21107309 | EFO_0004350 | ||
| ENSG00000142149 | rs2833610 | 21 | 32012873 | A | Type 2 diabetes | 21490949 | [1.09-1.24] | 1.17 | EFO_0001360 |
| ENSG00000142149 | rs2833579 | 21 | 31956217 | G | Periodontitis | 30218097 | [1.08-1.21] | 1.15 | EFO_0000649 |
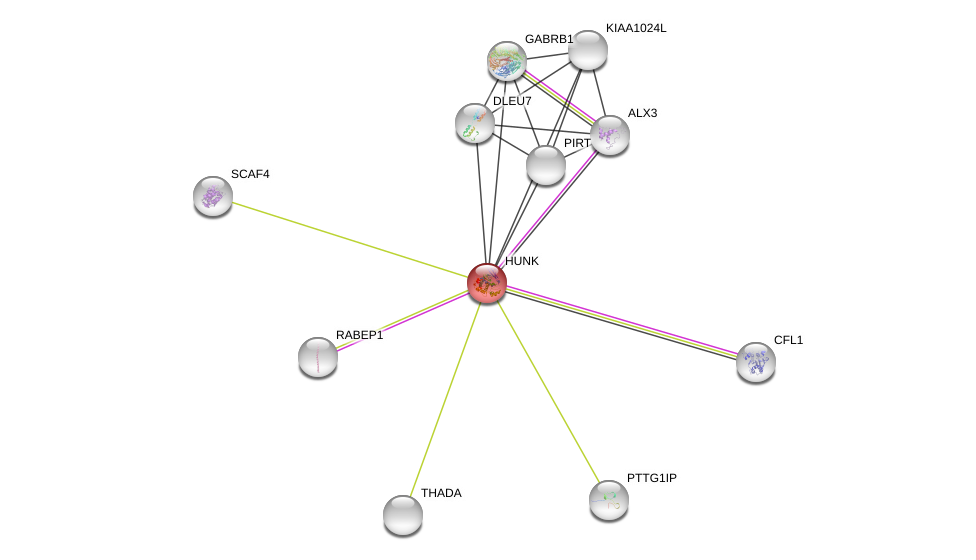
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000142149 | HUNK | 97 | 88.073 | ENSAMEG00000018210 | HUNK | 100 | 91.101 | Ailuropoda_melanoleuca |
| ENSG00000142149 | HUNK | 90 | 91.034 | ENSAPLG00000002063 | HUNK | 98 | 70.644 | Anas_platyrhynchos |
| ENSG00000142149 | HUNK | 90 | 88.276 | ENSACAG00000002743 | HUNK | 100 | 66.113 | Anolis_carolinensis |
| ENSG00000142149 | HUNK | 97 | 96.330 | ENSANAG00000026514 | - | 97 | 93.398 | Aotus_nancymaae |
| ENSG00000142149 | HUNK | 100 | 96.779 | ENSANAG00000018665 | - | 100 | 96.779 | Aotus_nancymaae |
| ENSG00000142149 | HUNK | 100 | 91.049 | ENSBTAG00000020762 | HUNK | 100 | 91.189 | Bos_taurus |
| ENSG00000142149 | HUNK | 100 | 97.199 | ENSCJAG00000045743 | HUNK | 100 | 97.199 | Callithrix_jacchus |
| ENSG00000142149 | HUNK | 97 | 88.991 | ENSCAFG00000008874 | HUNK | 98 | 90.635 | Canis_familiaris |
| ENSG00000142149 | HUNK | 100 | 89.103 | ENSCAFG00020026005 | HUNK | 100 | 90.345 | Canis_lupus_dingo |
| ENSG00000142149 | HUNK | 100 | 90.629 | ENSCHIG00000014504 | HUNK | 100 | 90.769 | Capra_hircus |
| ENSG00000142149 | HUNK | 97 | 87.156 | ENSTSYG00000005298 | HUNK | 100 | 82.380 | Carlito_syrichta |
| ENSG00000142149 | HUNK | 97 | 87.273 | ENSCAPG00000011721 | HUNK | 99 | 91.415 | Cavia_aperea |
| ENSG00000142149 | HUNK | 97 | 87.273 | ENSCPOG00000010546 | HUNK | 100 | 91.821 | Cavia_porcellus |
| ENSG00000142149 | HUNK | 100 | 96.639 | ENSCCAG00000028641 | HUNK | 100 | 96.639 | Cebus_capucinus |
| ENSG00000142149 | HUNK | 100 | 92.629 | ENSCATG00000028680 | HUNK | 100 | 92.629 | Cercocebus_atys |
| ENSG00000142149 | HUNK | 98 | 92.837 | ENSCLAG00000005512 | HUNK | 96 | 92.982 | Chinchilla_lanigera |
| ENSG00000142149 | HUNK | 97 | 98.165 | ENSCSAG00000009448 | HUNK | 100 | 98.997 | Chlorocebus_sabaeus |
| ENSG00000142149 | HUNK | 90 | 90.345 | ENSCPBG00000006541 | HUNK | 100 | 70.596 | Chrysemys_picta_bellii |
| ENSG00000142149 | HUNK | 97 | 100.000 | ENSCANG00000041035 | HUNK | 99 | 97.293 | Colobus_angolensis_palliatus |
| ENSG00000142149 | HUNK | 100 | 92.577 | ENSCGRG00001022573 | Hunk | 100 | 92.577 | Cricetulus_griseus_chok1gshd |
| ENSG00000142149 | HUNK | 97 | 88.991 | ENSCGRG00000013678 | Hunk | 100 | 92.185 | Cricetulus_griseus_crigri |
| ENSG00000142149 | HUNK | 100 | 91.469 | ENSDNOG00000039018 | HUNK | 100 | 92.168 | Dasypus_novemcinctus |
| ENSG00000142149 | HUNK | 90 | 93.793 | ENSDORG00000002143 | Hunk | 98 | 91.256 | Dipodomys_ordii |
| ENSG00000142149 | HUNK | 90 | 93.103 | ENSETEG00000011266 | HUNK | 100 | 88.712 | Echinops_telfairi |
| ENSG00000142149 | HUNK | 100 | 93.147 | ENSEASG00005007746 | HUNK | 100 | 92.308 | Equus_asinus_asinus |
| ENSG00000142149 | HUNK | 100 | 92.168 | ENSECAG00000007153 | HUNK | 100 | 92.308 | Equus_caballus |
| ENSG00000142149 | HUNK | 100 | 92.157 | ENSFCAG00000028905 | HUNK | 100 | 92.157 | Felis_catus |
| ENSG00000142149 | HUNK | 90 | 91.034 | ENSFALG00000008061 | HUNK | 100 | 73.225 | Ficedula_albicollis |
| ENSG00000142149 | HUNK | 97 | 88.182 | ENSFDAG00000009486 | HUNK | 100 | 90.127 | Fukomys_damarensis |
| ENSG00000142149 | HUNK | 90 | 82.069 | ENSGMOG00000015028 | HUNK | 93 | 74.788 | Gadus_morhua |
| ENSG00000142149 | HUNK | 90 | 91.034 | ENSGALG00000015879 | HUNK | 100 | 67.586 | Gallus_gallus |
| ENSG00000142149 | HUNK | 89 | 90.278 | ENSGAGG00000002645 | HUNK | 100 | 68.377 | Gopherus_agassizii |
| ENSG00000142149 | HUNK | 97 | 100.000 | ENSGGOG00000016651 | HUNK | 100 | 99.841 | Gorilla_gorilla |
| ENSG00000142149 | HUNK | 100 | 92.028 | ENSHGLG00000014832 | HUNK | 100 | 92.028 | Heterocephalus_glaber_female |
| ENSG00000142149 | HUNK | 100 | 92.028 | ENSHGLG00100007886 | HUNK | 100 | 92.028 | Heterocephalus_glaber_male |
| ENSG00000142149 | HUNK | 100 | 93.566 | ENSSTOG00000024744 | HUNK | 100 | 93.566 | Ictidomys_tridecemlineatus |
| ENSG00000142149 | HUNK | 97 | 88.991 | ENSJJAG00000014419 | Hunk | 98 | 93.142 | Jaculus_jaculus |
| ENSG00000142149 | HUNK | 83 | 86.567 | ENSLOCG00000009175 | HUNK | 93 | 49.639 | Lepisosteus_oculatus |
| ENSG00000142149 | HUNK | 95 | 89.051 | ENSLAFG00000004577 | HUNK | 100 | 89.051 | Loxodonta_africana |
| ENSG00000142149 | HUNK | 97 | 98.165 | ENSMFAG00000043007 | HUNK | 100 | 98.829 | Macaca_fascicularis |
| ENSG00000142149 | HUNK | 100 | 100.000 | ENSMMUG00000012917 | HUNK | 100 | 100.000 | Macaca_mulatta |
| ENSG00000142149 | HUNK | 99 | 97.335 | ENSMNEG00000044157 | HUNK | 92 | 97.335 | Macaca_nemestrina |
| ENSG00000142149 | HUNK | 97 | 98.165 | ENSMLEG00000038042 | HUNK | 100 | 99.043 | Mandrillus_leucophaeus |
| ENSG00000142149 | HUNK | 90 | 91.034 | ENSMGAG00000014513 | HUNK | 99 | 70.000 | Meleagris_gallopavo |
| ENSG00000142149 | HUNK | 100 | 87.500 | ENSMAUG00000018207 | Hunk | 82 | 95.354 | Mesocricetus_auratus |
| ENSG00000142149 | HUNK | 100 | 90.476 | ENSMOCG00000018032 | Hunk | 100 | 90.476 | Microtus_ochrogaster |
| ENSG00000142149 | HUNK | 100 | 92.577 | MGP_CAROLIEiJ_G0020945 | Hunk | 100 | 92.577 | Mus_caroli |
| ENSG00000142149 | HUNK | 100 | 92.437 | ENSMUSG00000053414 | Hunk | 100 | 92.437 | Mus_musculus |
| ENSG00000142149 | HUNK | 100 | 92.437 | MGP_PahariEiJ_G0016650 | Hunk | 100 | 91.317 | Mus_pahari |
| ENSG00000142149 | HUNK | 100 | 92.577 | MGP_SPRETEiJ_G0021851 | Hunk | 100 | 92.577 | Mus_spretus |
| ENSG00000142149 | HUNK | 97 | 86.239 | ENSMPUG00000001113 | HUNK | 100 | 90.468 | Mustela_putorius_furo |
| ENSG00000142149 | HUNK | 100 | 92.577 | ENSNGAG00000000191 | - | 100 | 92.577 | Nannospalax_galili |
| ENSG00000142149 | HUNK | 100 | 99.020 | ENSNLEG00000036238 | HUNK | 100 | 99.020 | Nomascus_leucogenys |
| ENSG00000142149 | HUNK | 100 | 88.039 | ENSOPRG00000009958 | HUNK | 100 | 87.761 | Ochotona_princeps |
| ENSG00000142149 | HUNK | 100 | 91.748 | ENSODEG00000002662 | HUNK | 100 | 91.748 | Octodon_degus |
| ENSG00000142149 | HUNK | 90 | 93.793 | ENSOCUG00000005096 | HUNK | 100 | 89.954 | Oryctolagus_cuniculus |
| ENSG00000142149 | HUNK | 100 | 91.597 | ENSOGAG00000010230 | HUNK | 100 | 91.597 | Otolemur_garnettii |
| ENSG00000142149 | HUNK | 95 | 91.259 | ENSOARG00000013875 | HUNK | 80 | 91.259 | Ovis_aries |
| ENSG00000142149 | HUNK | 97 | 100.000 | ENSPPAG00000032225 | HUNK | 98 | 97.866 | Pan_paniscus |
| ENSG00000142149 | HUNK | 100 | 92.157 | ENSPPRG00000022852 | HUNK | 100 | 92.157 | Panthera_pardus |
| ENSG00000142149 | HUNK | 97 | 88.182 | ENSPTIG00000021583 | HUNK | 99 | 90.938 | Panthera_tigris_altaica |
| ENSG00000142149 | HUNK | 100 | 99.720 | ENSPTRG00000013849 | HUNK | 100 | 99.720 | Pan_troglodytes |
| ENSG00000142149 | HUNK | 100 | 99.020 | ENSPANG00000022529 | HUNK | 100 | 99.020 | Papio_anubis |
| ENSG00000142149 | HUNK | 90 | 84.828 | ENSPKIG00000022484 | HUNK | 92 | 76.045 | Paramormyrops_kingsleyae |
| ENSG00000142149 | HUNK | 89 | 90.278 | ENSPSIG00000014236 | HUNK | 100 | 69.371 | Pelodiscus_sinensis |
| ENSG00000142149 | HUNK | 98 | 92.275 | ENSPEMG00000005678 | Hunk | 98 | 92.240 | Peromyscus_maniculatus_bairdii |
| ENSG00000142149 | HUNK | 90 | 90.345 | ENSPCIG00000014547 | HUNK | 100 | 75.829 | Phascolarctos_cinereus |
| ENSG00000142149 | HUNK | 97 | 100.000 | ENSPPYG00000011356 | HUNK | 100 | 93.417 | Pongo_abelii |
| ENSG00000142149 | HUNK | 90 | 91.034 | ENSPCAG00000001322 | HUNK | 100 | 79.787 | Procavia_capensis |
| ENSG00000142149 | HUNK | 100 | 95.098 | ENSPCOG00000007940 | HUNK | 100 | 95.098 | Propithecus_coquereli |
| ENSG00000142149 | HUNK | 100 | 90.756 | ENSPVAG00000005225 | HUNK | 100 | 90.756 | Pteropus_vampyrus |
| ENSG00000142149 | HUNK | 100 | 92.157 | ENSRNOG00000002092 | Hunk | 100 | 92.157 | Rattus_norvegicus |
| ENSG00000142149 | HUNK | 97 | 100.000 | ENSRBIG00000031500 | HUNK | 100 | 99.331 | Rhinopithecus_bieti |
| ENSG00000142149 | HUNK | 99 | 98.457 | ENSRROG00000037737 | HUNK | 99 | 98.457 | Rhinopithecus_roxellana |
| ENSG00000142149 | HUNK | 97 | 94.495 | ENSSBOG00000026563 | HUNK | 100 | 96.656 | Saimiri_boliviensis_boliviensis |
| ENSG00000142149 | HUNK | 90 | 90.345 | ENSSHAG00000009609 | HUNK | 99 | 76.341 | Sarcophilus_harrisii |
| ENSG00000142149 | HUNK | 90 | 86.207 | ENSSFOG00015006294 | HUNK | 90 | 52.087 | Scleropages_formosus |
| ENSG00000142149 | HUNK | 90 | 90.345 | ENSSPUG00000012338 | HUNK | 100 | 64.795 | Sphenodon_punctatus |
| ENSG00000142149 | HUNK | 97 | 87.156 | ENSSSCG00000029392 | HUNK | 92 | 94.030 | Sus_scrofa |
| ENSG00000142149 | HUNK | 90 | 91.034 | ENSTGUG00000013465 | HUNK | 100 | 72.117 | Taeniopygia_guttata |
| ENSG00000142149 | HUNK | 60 | 98.969 | ENSTTRG00000015604 | - | 64 | 99.200 | Tursiops_truncatus |
| ENSG00000142149 | HUNK | 97 | 88.991 | ENSUAMG00000027223 | HUNK | 97 | 91.228 | Ursus_americanus |
| ENSG00000142149 | HUNK | 97 | 88.991 | ENSUMAG00000017146 | HUNK | 98 | 91.011 | Ursus_maritimus |
| ENSG00000142149 | HUNK | 82 | 99.248 | ENSVPAG00000012399 | - | 68 | 99.379 | Vicugna_pacos |
| ENSG00000142149 | HUNK | 97 | 89.908 | ENSVVUG00000002915 | HUNK | 100 | 90.468 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004674 | protein serine/threonine kinase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 20133759. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0006468 | protein phosphorylation | 21873635. | IBA | Process |
| GO:0007165 | signal transduction | 10662544. | TAS | Process |
| GO:0007275 | multicellular organism development | 10662544. | TAS | Process |
| GO:0035556 | intracellular signal transduction | 21873635. | IBA | Process |