
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| ESCA | |||
| KIRC | |||
| PAAD | |||
| SKCM | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000402143 | Maelstrom | PF13017.6 | 1.1e-42 | 1 | 1 |
| ENSP00000482771 | Maelstrom | PF13017.6 | 1.6e-42 | 1 | 1 |
| ENSP00000356844 | Maelstrom | PF13017.6 | 1.9e-42 | 1 | 1 |
| ENSP00000356846 | Maelstrom | PF13017.6 | 2.3e-42 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000367870 | MAEL-201 | 1831 | - | ENSP00000356844 | 403 (aa) | - | Q96JY0 |
| ENST00000487826 | MAEL-204 | 416 | - | - | - (aa) | - | - |
| ENST00000367872 | MAEL-202 | 1908 | XM_006711583 | ENSP00000356846 | 434 (aa) | XP_006711646 | Q96JY0 |
| ENST00000491055 | MAEL-205 | 2355 | - | - | - (aa) | - | - |
| ENST00000622874 | MAEL-206 | 1700 | XM_011510068 | ENSP00000482771 | 378 (aa) | XP_011508370 | E9JVC4 |
| ENST00000447624 | MAEL-203 | 1013 | - | ENSP00000402143 | 316 (aa) | - | X6RGB1 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000143194 | Erythrocyte Indices | 7.5220000E-006 | - |
| ENSG00000143194 | Erythrocyte Indices | 5.8590000E-005 | - |
| ENSG00000143194 | Erythrocyte Indices | 5.8550000E-006 | - |
| ENSG00000143194 | Erythrocyte Count | 1.5470000E-005 | - |
| ENSG00000143194 | Erythrocyte Count | 7.0390000E-005 | - |
| ENSG00000143194 | Erythrocyte Count | 1.3720000E-005 | - |
| ENSG00000143194 | Platelet Function Tests | 1.1100000E-006 | - |
| ENSG00000143194 | Platelet Function Tests | 3.7800000E-007 | - |
| ENSG00000143194 | Platelet Function Tests | 9.0700000E-008 | - |
| ENSG00000143194 | Platelet Function Tests | 5.0100000E-006 | - |
| ENSG00000143194 | Platelet Function Tests | 4.3800000E-006 | - |
| ENSG00000143194 | Platelet Function Tests | 3.3593800E-005 | - |
| ENSG00000143194 | Platelet Function Tests | 1.0387300E-005 | - |
| ENSG00000143194 | Astigmatism | 3E-6 | 25367360 |
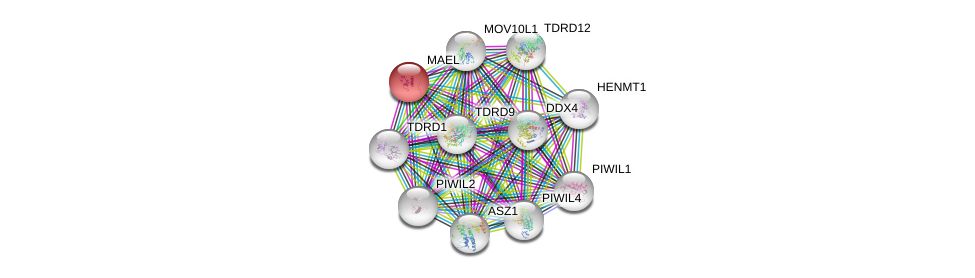
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000143194 | MAEL | 100 | 90.618 | ENSAMEG00000000018 | MAEL | 100 | 90.618 | Ailuropoda_melanoleuca |
| ENSG00000143194 | MAEL | 97 | 53.959 | ENSAPLG00000006696 | MAEL | 97 | 53.553 | Anas_platyrhynchos |
| ENSG00000143194 | MAEL | 73 | 53.879 | ENSACAG00000025602 | - | 100 | 49.822 | Anolis_carolinensis |
| ENSG00000143194 | MAEL | 100 | 55.848 | ENSACAG00000029327 | - | 84 | 55.025 | Anolis_carolinensis |
| ENSG00000143194 | MAEL | 100 | 93.671 | ENSANAG00000031520 | MAEL | 100 | 92.804 | Aotus_nancymaae |
| ENSG00000143194 | MAEL | 100 | 89.401 | ENSBTAG00000016550 | MAEL | 100 | 89.401 | Bos_taurus |
| ENSG00000143194 | MAEL | 100 | 94.620 | ENSCJAG00000017185 | MAEL | 100 | 94.293 | Callithrix_jacchus |
| ENSG00000143194 | MAEL | 100 | 92.627 | ENSCAFG00000015596 | MAEL | 100 | 93.103 | Canis_familiaris |
| ENSG00000143194 | MAEL | 100 | 92.627 | ENSCAFG00020022822 | MAEL | 100 | 92.627 | Canis_lupus_dingo |
| ENSG00000143194 | MAEL | 100 | 92.627 | ENSCHIG00000025356 | MAEL | 100 | 92.627 | Capra_hircus |
| ENSG00000143194 | MAEL | 100 | 91.798 | ENSTSYG00000003248 | MAEL | 100 | 89.425 | Carlito_syrichta |
| ENSG00000143194 | MAEL | 95 | 65.217 | ENSCAPG00000014130 | MAEL | 99 | 65.217 | Cavia_aperea |
| ENSG00000143194 | MAEL | 100 | 80.403 | ENSCPOG00000005046 | MAEL | 99 | 84.173 | Cavia_porcellus |
| ENSG00000143194 | MAEL | 100 | 95.392 | ENSCCAG00000033367 | MAEL | 100 | 95.392 | Cebus_capucinus |
| ENSG00000143194 | MAEL | 100 | 98.734 | ENSCATG00000036493 | MAEL | 100 | 97.767 | Cercocebus_atys |
| ENSG00000143194 | MAEL | 97 | 85.714 | ENSCLAG00000006862 | MAEL | 100 | 85.714 | Chinchilla_lanigera |
| ENSG00000143194 | MAEL | 100 | 97.701 | ENSCSAG00000010893 | MAEL | 100 | 97.701 | Chlorocebus_sabaeus |
| ENSG00000143194 | MAEL | 100 | 68.433 | ENSCHOG00000011901 | MAEL | 100 | 68.433 | Choloepus_hoffmanni |
| ENSG00000143194 | MAEL | 100 | 64.265 | ENSCPBG00000006089 | MAEL | 97 | 64.589 | Chrysemys_picta_bellii |
| ENSG00000143194 | MAEL | 100 | 98.734 | ENSCANG00000032047 | MAEL | 100 | 97.270 | Colobus_angolensis_palliatus |
| ENSG00000143194 | MAEL | 100 | 90.323 | ENSCGRG00001009188 | Mael | 100 | 90.323 | Cricetulus_griseus_chok1gshd |
| ENSG00000143194 | MAEL | 100 | 90.323 | ENSCGRG00000017037 | Mael | 100 | 90.323 | Cricetulus_griseus_crigri |
| ENSG00000143194 | MAEL | 100 | 90.847 | ENSDNOG00000016383 | MAEL | 100 | 90.847 | Dasypus_novemcinctus |
| ENSG00000143194 | MAEL | 100 | 88.940 | ENSDORG00000001149 | Mael | 100 | 88.940 | Dipodomys_ordii |
| ENSG00000143194 | MAEL | 82 | 85.874 | ENSETEG00000001331 | MAEL | 85 | 85.874 | Echinops_telfairi |
| ENSG00000143194 | MAEL | 100 | 90.553 | ENSEASG00005019126 | MAEL | 100 | 90.553 | Equus_asinus_asinus |
| ENSG00000143194 | MAEL | 100 | 90.553 | ENSECAG00000015077 | MAEL | 100 | 90.553 | Equus_caballus |
| ENSG00000143194 | MAEL | 98 | 80.328 | ENSEEUG00000014050 | MAEL | 100 | 80.328 | Erinaceus_europaeus |
| ENSG00000143194 | MAEL | 100 | 94.620 | ENSFCAG00000009285 | MAEL | 100 | 93.779 | Felis_catus |
| ENSG00000143194 | MAEL | 100 | 50.575 | ENSFALG00000001548 | MAEL | 98 | 50.372 | Ficedula_albicollis |
| ENSG00000143194 | MAEL | 95 | 87.591 | ENSFDAG00000012846 | MAEL | 100 | 87.591 | Fukomys_damarensis |
| ENSG00000143194 | MAEL | 79 | 60.558 | ENSGALG00000019211 | MAEL | 96 | 49.861 | Gallus_gallus |
| ENSG00000143194 | MAEL | 100 | 61.960 | ENSGAGG00000011274 | MAEL | 97 | 63.591 | Gopherus_agassizii |
| ENSG00000143194 | MAEL | 100 | 99.367 | ENSGGOG00000012560 | MAEL | 100 | 99.256 | Gorilla_gorilla |
| ENSG00000143194 | MAEL | 95 | 87.591 | ENSHGLG00000013438 | MAEL | 99 | 87.591 | Heterocephalus_glaber_female |
| ENSG00000143194 | MAEL | 99 | 84.985 | ENSHGLG00100014285 | MAEL | 99 | 81.156 | Heterocephalus_glaber_male |
| ENSG00000143194 | MAEL | 100 | 93.088 | ENSSTOG00000015434 | MAEL | 100 | 93.088 | Ictidomys_tridecemlineatus |
| ENSG00000143194 | MAEL | 100 | 85.714 | ENSJJAG00000011625 | Mael | 100 | 85.714 | Jaculus_jaculus |
| ENSG00000143194 | MAEL | 100 | 51.437 | ENSLACG00000008630 | MAEL | 99 | 53.086 | Latimeria_chalumnae |
| ENSG00000143194 | MAEL | 100 | 89.954 | ENSLAFG00000021470 | MAEL | 100 | 89.954 | Loxodonta_africana |
| ENSG00000143194 | MAEL | 100 | 99.051 | ENSMFAG00000040593 | MAEL | 100 | 98.511 | Macaca_fascicularis |
| ENSG00000143194 | MAEL | 100 | 98.413 | ENSMMUG00000023463 | MAEL | 100 | 98.413 | Macaca_mulatta |
| ENSG00000143194 | MAEL | 100 | 99.051 | ENSMNEG00000042870 | MAEL | 100 | 98.263 | Macaca_nemestrina |
| ENSG00000143194 | MAEL | 100 | 98.734 | ENSMLEG00000034260 | MAEL | 100 | 98.015 | Mandrillus_leucophaeus |
| ENSG00000143194 | MAEL | 100 | 51.429 | ENSMGAG00000014437 | MAEL | 95 | 50.000 | Meleagris_gallopavo |
| ENSG00000143194 | MAEL | 100 | 89.862 | ENSMAUG00000021599 | Mael | 100 | 89.862 | Mesocricetus_auratus |
| ENSG00000143194 | MAEL | 100 | 95.886 | ENSMICG00000034747 | MAEL | 100 | 94.931 | Microcebus_murinus |
| ENSG00000143194 | MAEL | 100 | 88.249 | ENSMOCG00000004042 | Mael | 100 | 87.558 | Microtus_ochrogaster |
| ENSG00000143194 | MAEL | 100 | 78.082 | ENSMODG00000002482 | MAEL | 100 | 78.082 | Monodelphis_domestica |
| ENSG00000143194 | MAEL | 100 | 89.862 | MGP_CAROLIEiJ_G0014814 | Mael | 100 | 89.862 | Mus_caroli |
| ENSG00000143194 | MAEL | 100 | 89.862 | ENSMUSG00000040629 | Mael | 100 | 89.862 | Mus_musculus |
| ENSG00000143194 | MAEL | 100 | 89.862 | MGP_PahariEiJ_G0028051 | Mael | 100 | 89.862 | Mus_pahari |
| ENSG00000143194 | MAEL | 100 | 89.862 | MGP_SPRETEiJ_G0015621 | Mael | 100 | 89.862 | Mus_spretus |
| ENSG00000143194 | MAEL | 100 | 91.475 | ENSMPUG00000015684 | MAEL | 100 | 91.475 | Mustela_putorius_furo |
| ENSG00000143194 | MAEL | 100 | 87.414 | ENSMLUG00000004095 | MAEL | 100 | 87.414 | Myotis_lucifugus |
| ENSG00000143194 | MAEL | 99 | 89.277 | ENSNGAG00000013179 | Mael | 99 | 89.277 | Nannospalax_galili |
| ENSG00000143194 | MAEL | 100 | 98.418 | ENSNLEG00000014771 | MAEL | 100 | 98.263 | Nomascus_leucogenys |
| ENSG00000143194 | MAEL | 100 | 68.433 | ENSMEUG00000001673 | - | 100 | 68.433 | Notamacropus_eugenii |
| ENSG00000143194 | MAEL | 100 | 80.000 | ENSOPRG00000005269 | MAEL | 100 | 80.000 | Ochotona_princeps |
| ENSG00000143194 | MAEL | 95 | 84.951 | ENSODEG00000002276 | MAEL | 99 | 84.951 | Octodon_degus |
| ENSG00000143194 | MAEL | 79 | 65.882 | ENSOANG00000003874 | - | 98 | 55.840 | Ornithorhynchus_anatinus |
| ENSG00000143194 | MAEL | 100 | 89.885 | ENSOCUG00000005848 | MAEL | 100 | 89.885 | Oryctolagus_cuniculus |
| ENSG00000143194 | MAEL | 100 | 94.037 | ENSOGAG00000007627 | MAEL | 100 | 94.037 | Otolemur_garnettii |
| ENSG00000143194 | MAEL | 100 | 92.220 | ENSOARG00000011888 | MAEL | 100 | 92.220 | Ovis_aries |
| ENSG00000143194 | MAEL | 100 | 99.256 | ENSPPAG00000042153 | MAEL | 100 | 99.256 | Pan_paniscus |
| ENSG00000143194 | MAEL | 100 | 94.937 | ENSPPRG00000003002 | MAEL | 100 | 93.548 | Panthera_pardus |
| ENSG00000143194 | MAEL | 100 | 93.915 | ENSPTIG00000011034 | MAEL | 93 | 93.878 | Panthera_tigris_altaica |
| ENSG00000143194 | MAEL | 100 | 99.256 | ENSPTRG00000001629 | MAEL | 100 | 99.256 | Pan_troglodytes |
| ENSG00000143194 | MAEL | 100 | 98.734 | ENSPANG00000000174 | MAEL | 100 | 98.263 | Papio_anubis |
| ENSG00000143194 | MAEL | 100 | 60.684 | ENSPSIG00000009813 | MAEL | 97 | 62.283 | Pelodiscus_sinensis |
| ENSG00000143194 | MAEL | 74 | 77.778 | ENSPCIG00000026830 | - | 96 | 77.778 | Phascolarctos_cinereus |
| ENSG00000143194 | MAEL | 100 | 76.256 | ENSPCIG00000030034 | - | 100 | 76.256 | Phascolarctos_cinereus |
| ENSG00000143194 | MAEL | 100 | 98.677 | ENSPPYG00000000555 | MAEL | 100 | 98.618 | Pongo_abelii |
| ENSG00000143194 | MAEL | 95 | 72.145 | ENSPCAG00000013028 | MAEL | 100 | 72.145 | Procavia_capensis |
| ENSG00000143194 | MAEL | 100 | 94.620 | ENSPCOG00000007295 | MAEL | 100 | 93.318 | Propithecus_coquereli |
| ENSG00000143194 | MAEL | 100 | 85.591 | ENSPVAG00000013294 | MAEL | 100 | 83.180 | Pteropus_vampyrus |
| ENSG00000143194 | MAEL | 98 | 90.610 | ENSRNOG00000003790 | Mael | 97 | 90.610 | Rattus_norvegicus |
| ENSG00000143194 | MAEL | 100 | 99.051 | ENSRBIG00000038763 | MAEL | 100 | 98.263 | Rhinopithecus_bieti |
| ENSG00000143194 | MAEL | 100 | 99.051 | ENSRROG00000039365 | MAEL | 100 | 98.263 | Rhinopithecus_roxellana |
| ENSG00000143194 | MAEL | 100 | 94.937 | ENSSBOG00000027193 | MAEL | 100 | 94.931 | Saimiri_boliviensis_boliviensis |
| ENSG00000143194 | MAEL | 100 | 72.585 | ENSSHAG00000018323 | MAEL | 100 | 73.418 | Sarcophilus_harrisii |
| ENSG00000143194 | MAEL | 100 | 61.404 | ENSSPUG00000018576 | MAEL | 96 | 60.101 | Sphenodon_punctatus |
| ENSG00000143194 | MAEL | 100 | 91.705 | ENSSSCG00000006316 | MAEL | 100 | 91.705 | Sus_scrofa |
| ENSG00000143194 | MAEL | 100 | 48.547 | ENSTGUG00000013511 | MAEL | 100 | 50.700 | Taeniopygia_guttata |
| ENSG00000143194 | MAEL | 77 | 82.909 | ENSTBEG00000003322 | MAEL | 85 | 75.068 | Tupaia_belangeri |
| ENSG00000143194 | MAEL | 100 | 83.598 | ENSTTRG00000006132 | MAEL | 100 | 84.670 | Tursiops_truncatus |
| ENSG00000143194 | MAEL | 100 | 91.705 | ENSUAMG00000018439 | MAEL | 100 | 91.705 | Ursus_americanus |
| ENSG00000143194 | MAEL | 100 | 91.270 | ENSUMAG00000016528 | MAEL | 99 | 91.408 | Ursus_maritimus |
| ENSG00000143194 | MAEL | 84 | 91.880 | ENSVPAG00000004937 | MAEL | 84 | 91.880 | Vicugna_pacos |
| ENSG00000143194 | MAEL | 100 | 92.593 | ENSVVUG00000026348 | MAEL | 100 | 92.593 | Vulpes_vulpes |
| ENSG00000143194 | MAEL | 88 | 47.670 | ENSXETG00000027834 | mael | 97 | 42.632 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0000785 | chromatin | - | IEA | Component |
| GO:0000902 | cell morphogenesis | - | IEA | Process |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | ISM | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | NAS | Function |
| GO:0001741 | XY body | - | IEA | Component |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | - | ISS | Component |
| GO:0005737 | cytoplasm | - | ISS | Component |
| GO:0007129 | synapsis | - | IEA | Process |
| GO:0007140 | male meiotic nuclear division | 21873635. | IBA | Process |
| GO:0007275 | multicellular organism development | - | IEA | Process |
| GO:0007283 | spermatogenesis | 21873635. | IBA | Process |
| GO:0007283 | spermatogenesis | - | ISS | Process |
| GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | - | IEA | Process |
| GO:0009566 | fertilization | - | IEA | Process |
| GO:0030154 | cell differentiation | - | IEA | Process |
| GO:0030849 | autosome | - | IEA | Component |
| GO:0031047 | gene silencing by RNA | - | ISS | Process |
| GO:0033391 | chromatoid body | - | IEA | Component |
| GO:0034587 | piRNA metabolic process | 21873635. | IBA | Process |
| GO:0034587 | piRNA metabolic process | - | ISS | Process |
| GO:0043046 | DNA methylation involved in gamete generation | - | ISS | Process |
| GO:0043066 | negative regulation of apoptotic process | - | IEA | Process |
| GO:0043186 | P granule | 21873635. | IBA | Component |
| GO:0043186 | P granule | - | ISS | Component |
| GO:0043565 | sequence-specific DNA binding | 21873635. | IBA | Function |
| GO:0045892 | negative regulation of transcription, DNA-templated | 21873635. | IBA | Process |
| GO:0046620 | regulation of organ growth | - | IEA | Process |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0060964 | regulation of gene silencing by miRNA | - | IEA | Process |
| GO:0071547 | piP-body | - | ISS | Component |