
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LAML | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| SKCM | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| LAML | ||
| GBM | ||
| LIHC | ||
| LGG |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000488324 | PPP1R18-204 | 1328 | - | - | - (aa) | - | - |
| ENST00000615527 | PPP1R18-205 | 4597 | - | ENSP00000480270 | 613 (aa) | - | Q6NYC8 |
| ENST00000274853 | PPP1R18-201 | 4599 | - | ENSP00000274853 | 613 (aa) | - | Q6NYC8 |
| ENST00000399199 | PPP1R18-202 | 2853 | - | ENSP00000382150 | 613 (aa) | - | Q6NYC8 |
| ENST00000467662 | PPP1R18-203 | 1156 | - | - | - (aa) | - | - |
| ENST00000615892 | PPP1R18-206 | 1448 | - | ENSP00000482578 | 165 (aa) | - | Q6NYC8 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000146112 | rs116295105 | 6 | 30685004 | T | Lung cancer | 28604730 | [1.136875795-1.225357708] | 1.1802878 | EFO_0001071 |
| ENSG00000146112 | rs9262142 | 6 | 30682249 | A | Breast cancer | 29059683 | [0.028-0.069] unit decrease | 0.0489 | EFO_0000305 |
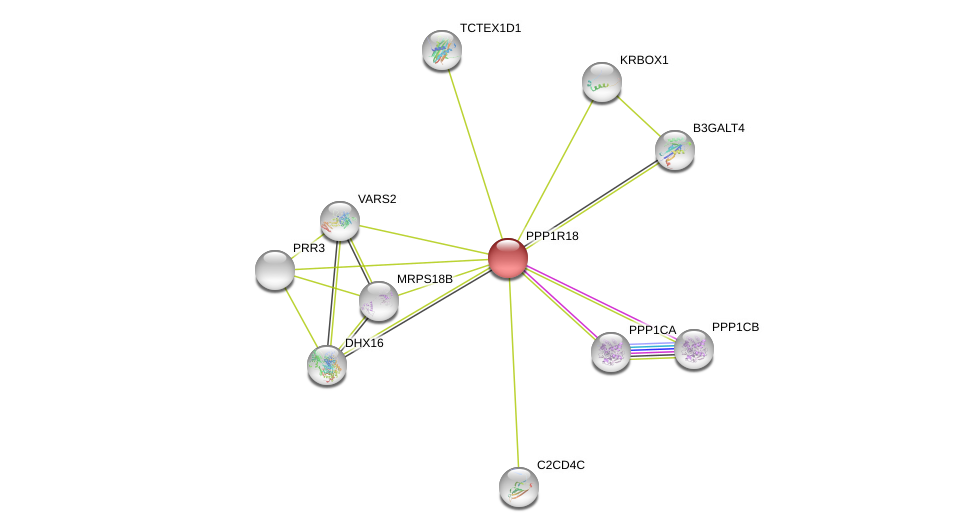
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003779 | actin binding | - | IEA | Function |
| GO:0005515 | protein binding | 21516116.22321011.25416956.25593058.28330616. | IPI | Function |
| GO:0005737 | cytoplasm | - | IEA | Component |
| GO:0005856 | cytoskeleton | - | IEA | Component |
| GO:0019902 | phosphatase binding | - | IEA | Function |