
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| LUAD |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| SARC | ||
| ACC | ||
| LUSC | ||
| KIRP | ||
| PAAD | ||
| CESC | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| UVM | ||
| OV |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 21346410 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000550475 | PRIM2-206 | 375 | - | ENSP00000485395 | 93 (aa) | - | A0A096LP51 |
| ENST00000470638 | PRIM2-204 | 486 | - | - | - (aa) | - | - |
| ENST00000615550 | PRIM2-207 | 2308 | XM_017011004 | ENSP00000484105 | 509 (aa) | XP_016866493 | P49643 |
| ENST00000370687 | PRIM2-202 | 909 | - | ENSP00000483201 | 158 (aa) | - | P49643 |
| ENST00000274891 | PRIM2-201 | 864 | - | ENSP00000485304 | 158 (aa) | - | P49643 |
| ENST00000490313 | PRIM2-205 | 410 | - | - | - (aa) | - | - |
| ENST00000419977 | PRIM2-203 | 2197 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03030 | DNA replication | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000146143 | Arteries | 6.7250000E-005 | - |
| ENSG00000146143 | Ankle Brachial Index | 3.4050031E-005 | - |
| ENSG00000146143 | Ankle Brachial Index | 8.2186358E-005 | - |
| ENSG00000146143 | Ankle Brachial Index | 3.7276687E-005 | - |
| ENSG00000146143 | Ankle Brachial Index | 3.3854710E-005 | - |
| ENSG00000146143 | Ankle Brachial Index | 1.9461675E-005 | - |
| ENSG00000146143 | 25-Hydroxyvitamin D 2 | 4.3180000E-005 | - |
| ENSG00000146143 | Longevity | 4E-6 | 27015805 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000146143 | rs3800007 | 6 | 57372179 | ? | Parental extreme longevity (95 years and older) | 27015805 | [0.0071-0.0177] unit decrease | 0.01239 | EFO_0007796 |
| ENSG00000146143 | rs2013268 | 6 | 57404607 | A | Red vs. brown/black hair color | 30531825 | NR | 288.8 | EFO_0003924 |
| ENSG00000146143 | rs9476035 | 6 | 57494269 | ? | Lung function (FEV1/FVC) | 30595370 | EFO_0004713 |
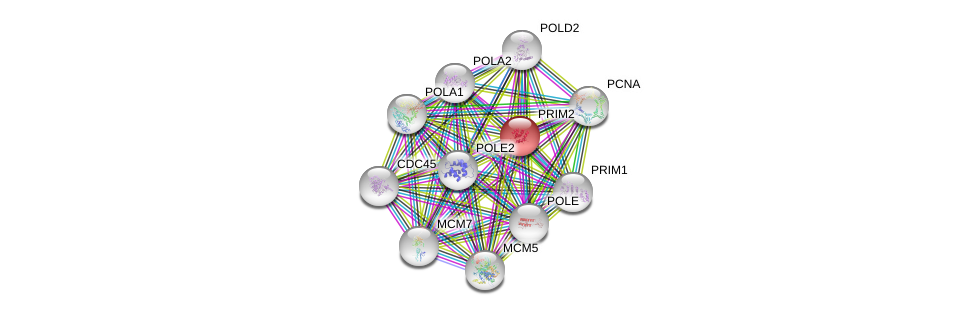
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000082 | G1/S transition of mitotic cell cycle | - | TAS | Process |
| GO:0003697 | single-stranded DNA binding | 21873635. | IBA | Function |
| GO:0003896 | DNA primase activity | 21873635. | IBA | Function |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005658 | alpha DNA polymerase:primase complex | 21873635. | IBA | Component |
| GO:0006269 | DNA replication, synthesis of RNA primer | - | IEA | Process |
| GO:0006270 | DNA replication initiation | - | TAS | Process |
| GO:0032201 | telomere maintenance via semi-conservative replication | - | TAS | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0051539 | 4 iron, 4 sulfur cluster binding | - | IEA | Function |