
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| BLCA | |||
| ESCA | |||
| HNSC | |||
| HNSC | |||
| LAML | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| SARC | ||
| ACC | ||
| LUSC | ||
| PAAD | ||
| BLCA | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| UVM |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 24425787 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000374773 | PARD3-205 | 3420 | - | ENSP00000363905 | 1001 (aa) | - | Q5VWU8 |
| ENST00000374776 | PARD3-206 | 3389 | - | ENSP00000363908 | 988 (aa) | - | Q8TEW0 |
| ENST00000374788 | PARD3-207 | 5996 | - | ENSP00000363920 | 1353 (aa) | - | Q8TEW0 |
| ENST00000374789 | PARD3-208 | 6005 | - | ENSP00000363921 | 1356 (aa) | - | Q8TEW0 |
| ENST00000374768 | PARD3-204 | 799 | - | ENSP00000363900 | 179 (aa) | - | B1AP52 |
| ENST00000466092 | PARD3-211 | 806 | - | - | - (aa) | - | - |
| ENST00000350537 | PARD3-203 | 5867 | - | ENSP00000311986 | 1310 (aa) | - | Q8TEW0 |
| ENST00000545693 | PARD3-214 | 5962 | - | ENSP00000443147 | 1340 (aa) | - | Q8TEW0 |
| ENST00000374790 | PARD3-209 | 5825 | - | ENSP00000363922 | 1296 (aa) | - | Q5VWV2 |
| ENST00000544292 | PARD3-212 | 2475 | - | ENSP00000444429 | 747 (aa) | - | F5GZI3 |
| ENST00000374794 | PARD3-210 | 5669 | - | ENSP00000363926 | 1244 (aa) | - | Q8TEW0 |
| ENST00000545260 | PARD3-213 | 5740 | - | ENSP00000440857 | 1266 (aa) | - | Q8TEW0 |
| ENST00000340077 | PARD3-201 | 3518 | - | ENSP00000341844 | 1031 (aa) | - | Q8TEW0 |
| ENST00000346874 | PARD3-202 | 5894 | - | ENSP00000340591 | 1319 (aa) | - | Q8TEW0 |
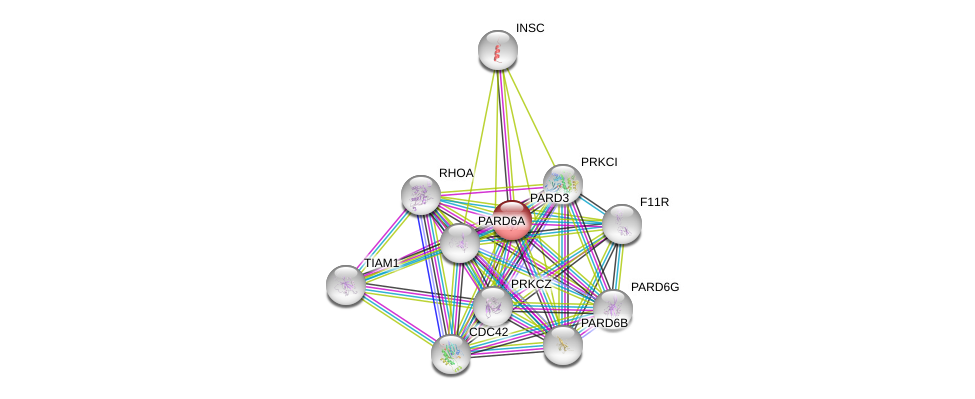
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | KEGG |
| hsa04062 | Chemokine signaling pathway | KEGG |
| hsa04080 | Neuroactive ligand-receptor interaction | KEGG |
| hsa04144 | Endocytosis | KEGG |
| hsa04360 | Axon guidance | KEGG |
| hsa04390 | Hippo signaling pathway | KEGG |
| hsa04520 | Adherens junction | KEGG |
| hsa04530 | Tight junction | KEGG |
| hsa05165 | Human papillomavirus infection | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000148498 | Hip | 1.04812188133394E-5 | 17903296 |
| ENSG00000148498 | Hip | 1.74693336596476E-5 | 17903296 |
| ENSG00000148498 | Intelligence Tests | 1.7766400E-005 | - |
| ENSG00000148498 | Child Development Disorders, Pervasive | 1.6277500E-005 | - |
| ENSG00000148498 | Precursor Cell Lymphoblastic Leukemia-Lymphoma | 9E-6 | 19684603 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000148498 | rs563507 | 10 | 34529060 | A | Acute lymphoblastic leukemia (childhood) | 19684603 | [1.40-2.70] | 2 | EFO_0000220 |
| ENSG00000148498 | rs2197012 | 10 | 34294707 | A | Educational attainment (MTAG) | 30038396 | [0.0069-0.0119] unit increase | 0.0094 | EFO_0004784 |
| ENSG00000148498 | rs1765174 | 10 | 34279104 | C | Highest math class taken | 30038396 | [0.0097-0.0187] unit increase | 0.0142 | EFO_0004875 |
| ENSG00000148498 | rs641162 | 10 | 34277992 | A | Self-reported math ability (MTAG) | 30038396 | [0.008-0.015] unit decrease | 0.0115 | EFO_0004875 |
| ENSG00000148498 | rs641162 | 10 | 34277992 | A | Highest math class taken (MTAG) | 30038396 | [0.0094-0.016] unit decrease | 0.0127 | EFO_0004875 |
| ENSG00000148498 | rs142712198 | 10 | 34449119 | ? | General cognitive ability | 29844566 | z-score increase | 4.533 | EFO_0004337 |
| ENSG00000148498 | rs73267375 | 10 | 34600928 | ? | General cognitive ability | 29844566 | z-score decrease | 4.47 | EFO_0004337 |
| ENSG00000148498 | rs2007655 | 10 | 34297761 | T | Educational attainment (years of education) | 30038396 | [0.0059-0.0113] unit increase | 0.0086 | EFO_0004784 |
| ENSG00000148498 | rs647758 | 10 | 34201926 | ? | Lung function (FVC) | 30595370 | EFO_0004312 | ||
| ENSG00000148498 | rs3004929 | 10 | 34125937 | ? | Body mass index | 30595370 | EFO_0004340 | ||
| ENSG00000148498 | rs1274475 | 10 | 34191654 | ? | Lung function (FEV1/FVC) | 30595370 | EFO_0004713 | ||
| ENSG00000148498 | rs1274475 | 10 | 34191654 | A | Lung function (FEV1/FVC) | 30804560 | [0.012-0.022] unit increase | 0.0168 | EFO_0004713 |
| ENSG00000148498 | rs138016380 | 10 | 34160538 | C | Corneal astigmatism | 30306274 | [0.026-0.062] dioptres decrease | 0.044 | EFO_1002040 |
| ENSG00000148498 | rs11009904 | 10 | 34735416 | ? | Heel bone mineral density | 30595370 | EFO_0009270 |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000226 | microtubule cytoskeleton organization | 21873635. | IBA | Process |
| GO:0005515 | protein binding | 10934474.11257119.14676191.15161933.15254234.17057644.17979178.18838552.19893486.22579248.22653443.25241761.26496610.26766442. | IPI | Function |
| GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | - | ISS | Function |
| GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding | - | ISS | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005856 | cytoskeleton | - | IEA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005911 | cell-cell junction | 20332120. | IDA | Component |
| GO:0005911 | cell-cell junction | - | ISS | Component |
| GO:0005912 | adherens junction | 21873635. | IBA | Component |
| GO:0005913 | cell-cell adherens junction | 21873635. | IBA | Component |
| GO:0005923 | bicellular tight junction | 14676191.22006950. | IDA | Component |
| GO:0005923 | bicellular tight junction | - | ISS | Component |
| GO:0005938 | cell cortex | 21873635. | IBA | Component |
| GO:0006612 | protein targeting to membrane | - | ISS | Process |
| GO:0007049 | cell cycle | - | IEA | Process |
| GO:0007155 | cell adhesion | 21873635. | IBA | Process |
| GO:0007163 | establishment or maintenance of cell polarity | 14676191. | TAS | Process |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | - | TAS | Process |
| GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 11260256. | TAS | Process |
| GO:0007409 | axonogenesis | 14676191. | TAS | Process |
| GO:0008104 | protein localization | 21873635. | IBA | Process |
| GO:0008356 | asymmetric cell division | 10934474. | TAS | Process |
| GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | - | ISS | Process |
| GO:0012505 | endomembrane system | - | IEA | Component |
| GO:0016324 | apical plasma membrane | 21873635. | IBA | Component |
| GO:0019903 | protein phosphatase binding | - | IEA | Function |
| GO:0022011 | myelination in peripheral nervous system | - | ISS | Process |
| GO:0030010 | establishment of cell polarity | 21873635. | IBA | Process |
| GO:0030054 | cell junction | - | IDA | Component |
| GO:0030054 | cell junction | - | TAS | Component |
| GO:0031643 | positive regulation of myelination | - | ISS | Process |
| GO:0032266 | phosphatidylinositol-3-phosphate binding | - | ISS | Function |
| GO:0032991 | protein-containing complex | - | IEA | Component |
| GO:0033269 | internode region of axon | - | ISS | Component |
| GO:0035091 | phosphatidylinositol binding | 21873635. | IBA | Function |
| GO:0042802 | identical protein binding | - | IEA | Function |
| GO:0043025 | neuronal cell body | - | IEA | Component |
| GO:0043296 | apical junction complex | 21873635. | IBA | Component |
| GO:0044295 | axonal growth cone | - | IEA | Component |
| GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 21873635. | IBA | Process |
| GO:0051660 | establishment of centrosome localization | 21873635. | IBA | Process |
| GO:0060341 | regulation of cellular localization | - | IEA | Process |
| GO:0065003 | protein-containing complex assembly | 10934474. | TAS | Process |
| GO:0070830 | bicellular tight junction assembly | - | ISS | Process |
| GO:0070830 | bicellular tight junction assembly | - | TAS | Process |
| GO:0090162 | establishment of epithelial cell polarity | - | ISS | Process |