
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 26700459 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| CESC | |||
| CHOL | |||
| ESCA | |||
| KIRP | |||
| LGG | |||
| LIHC | |||
| LUAD | |||
| PRAD | |||
| READ | |||
| SARC | |||
| STAD | |||
| TGCT | |||
| UCS | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| SARC | ||
| ACC | ||
| HNSC | ||
| TGCT | ||
| KIRP | ||
| COAD | ||
| BLCA | ||
| CESC | ||
| READ | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| THCA | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000617285 | HYOU1-220 | 4529 | XM_005271392 | ENSP00000480150 | 999 (aa) | XP_005271449 | Q9Y4L1 |
| ENST00000531682 | HYOU1-208 | 319 | - | - | - (aa) | - | - |
| ENST00000532519 | HYOU1-212 | 2162 | - | ENSP00000467866 | 678 (aa) | - | K7EQK2 |
| ENST00000529174 | HYOU1-205 | 549 | - | - | - (aa) | - | - |
| ENST00000543287 | HYOU1-215 | 2464 | - | ENSP00000442727 | 38 (aa) | - | J3KSR2 |
| ENST00000527038 | HYOU1-203 | 629 | - | ENSP00000463742 | 149 (aa) | - | J3QQH7 |
| ENST00000526656 | HYOU1-202 | 862 | - | ENSP00000463318 | 211 (aa) | - | J3QL06 |
| ENST00000610597 | HYOU1-216 | 491 | - | ENSP00000484622 | 92 (aa) | - | A0A087X214 |
| ENST00000527738 | HYOU1-204 | 809 | - | - | - (aa) | - | - |
| ENST00000621959 | HYOU1-221 | 3358 | - | ENSP00000484186 | 937 (aa) | - | A0A087X054 |
| ENST00000533381 | HYOU1-213 | 694 | - | - | - (aa) | - | - |
| ENST00000531968 | HYOU1-210 | 550 | - | ENSP00000464026 | 38 (aa) | - | J3KSR2 |
| ENST00000530467 | HYOU1-206 | 552 | - | ENSP00000479845 | 102 (aa) | - | A0A087WW13 |
| ENST00000531694 | HYOU1-209 | 2357 | - | - | - (aa) | - | - |
| ENST00000532421 | HYOU1-211 | 582 | - | ENSP00000463515 | 154 (aa) | - | J3QLE9 |
| ENST00000612687 | HYOU1-217 | 4360 | - | ENSP00000483106 | 937 (aa) | - | A0A087X054 |
| ENST00000534233 | HYOU1-214 | 711 | - | ENSP00000462951 | 224 (aa) | - | J3KTF1 |
| ENST00000614711 | HYOU1-219 | 3057 | - | ENSP00000480248 | 677 (aa) | - | A0A087WWI4 |
| ENST00000614668 | HYOU1-218 | 1569 | - | ENSP00000482199 | 147 (aa) | - | Q9BST8 |
| ENST00000526354 | HYOU1-201 | 568 | - | - | - (aa) | - | - |
| ENST00000622474 | HYOU1-222 | 549 | - | - | - (aa) | - | - |
| ENST00000530473 | HYOU1-207 | 2023 | - | ENSP00000431874 | 656 (aa) | - | E9PJ21 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04141 | Protein processing in endoplasmic reticulum | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000149428 | rs147134271 | 11 | 119053427 | G | Post bronchodilator FEV1/FVC ratio | 26634245 | unit increase | 0.231 | GO_0097366|EFO_0004713 |
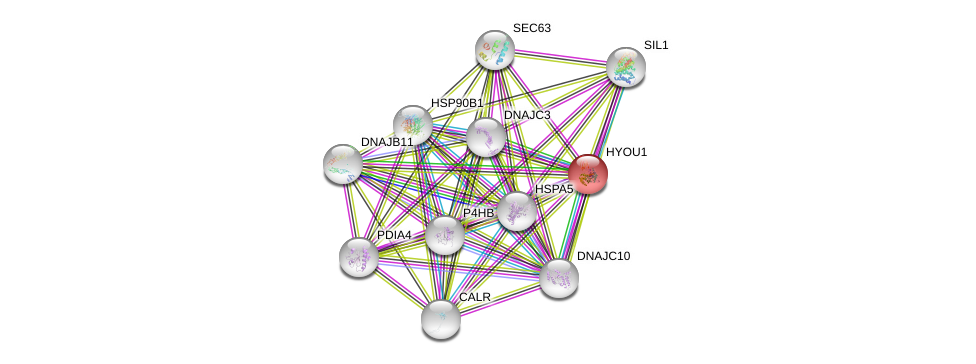
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000149428 | HYOU1 | 100 | 93.598 | ENSMUSG00000032115 | Hyou1 | 100 | 92.092 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002931 | response to ischemia | - | ISS | Process |
| GO:0005515 | protein binding | 26496610.30021884. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005783 | endoplasmic reticulum | 21873635. | IBA | Component |
| GO:0005783 | endoplasmic reticulum | 9020069.10837345. | NAS | Component |
| GO:0005788 | endoplasmic reticulum lumen | - | TAS | Component |
| GO:0005790 | smooth endoplasmic reticulum | - | ISS | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0006888 | ER to Golgi vesicle-mediated transport | 10837345. | IDA | Process |
| GO:0006898 | receptor-mediated endocytosis | - | TAS | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0034663 | endoplasmic reticulum chaperone complex | 21873635. | IBA | Component |
| GO:0034976 | response to endoplasmic reticulum stress | 9020069. | IEP | Process |
| GO:0036498 | IRE1-mediated unfolded protein response | - | TAS | Process |
| GO:0051087 | chaperone binding | 10837345. | ISS | Function |
| GO:0070062 | extracellular exosome | 19199708. | HDA | Component |
| GO:0071456 | cellular response to hypoxia | 21873635. | IBA | Process |
| GO:0071456 | cellular response to hypoxia | 10837345. | IDA | Process |
| GO:0071456 | cellular response to hypoxia | 9020069. | IEP | Process |
| GO:0071682 | endocytic vesicle lumen | - | TAS | Component |
| GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 21873635. | IBA | Process |
| GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 10037731. | IDA | Process |
| GO:1903382 | negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | - | IEA | Process |