
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 14533006 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| GBM | |||
| KIRC | |||
| LGG | |||
| MESO | |||
| PAAD | |||
| SKCM | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| HNSC | ||
| SKCM | ||
| PAAD | ||
| BLCA | ||
| UCEC | ||
| GBM | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000350265 | Endonuclease_NS | PF01223.23 | 8e-06 | 1 | 1 |
| ENSP00000406261 | Endonuclease_NS | PF01223.23 | 8e-06 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000358229 | ENPP3-202 | 3026 | - | ENSP00000350964 | 663 (aa) | - | F8W6H5 |
| ENST00000494023 | ENPP3-207 | 632 | - | - | - (aa) | - | - |
| ENST00000357639 | ENPP3-201 | 3162 | XM_011535897 | ENSP00000350265 | 875 (aa) | XP_011534199 | O14638 |
| ENST00000423831 | ENPP3-204 | 598 | - | ENSP00000408642 | 53 (aa) | - | E9PDB4 |
| ENST00000470930 | ENPP3-206 | 550 | - | - | - (aa) | - | - |
| ENST00000427707 | ENPP3-205 | 1536 | - | ENSP00000415589 | 53 (aa) | - | E7ETI7 |
| ENST00000414305 | ENPP3-203 | 3418 | - | ENSP00000406261 | 875 (aa) | - | O14638 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000154269 | rs12199015 | 6 | 131737807 | ? | Response to fenofibrate (adiponectin levels) | 23149075 | [0.051-0.129] ng/dL increase | 0.09 | GO_1901557 |
| ENSG00000154269 | rs17601580 | 6 | 131740280 | ? | Body mass index (change over time) in lung cancer | 28044437 | [0.069-0.155] unit decrease | 0.112 | EFO_0001071|EFO_0005937 |
| ENSG00000154269 | rs7766061 | 6 | 131645893 | ? | Reaction time | 29844566 | [0.0047-0.0116] unit decrease | 0.0081433 | EFO_0008393 |
| ENSG00000154269 | rs9321309 | 6 | 131726105 | ? | Adolescent idiopathic scoliosis | 30019117 | EFO_0005423 |
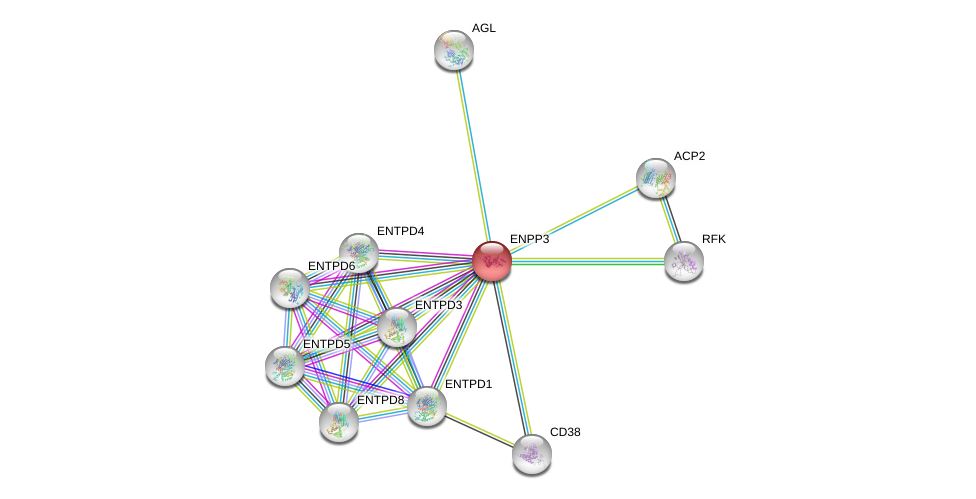
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000154269 | ENPP3 | 100 | 83.580 | ENSAMEG00000010324 | ENPP3 | 100 | 83.580 | Ailuropoda_melanoleuca |
| ENSG00000154269 | ENPP3 | 99 | 66.858 | ENSAPLG00000009807 | ENPP3 | 99 | 66.628 | Anas_platyrhynchos |
| ENSG00000154269 | ENPP3 | 97 | 67.488 | ENSACAG00000001394 | ENPP3 | 97 | 67.488 | Anolis_carolinensis |
| ENSG00000154269 | ENPP3 | 100 | 94.171 | ENSANAG00000029995 | ENPP3 | 100 | 94.171 | Aotus_nancymaae |
| ENSG00000154269 | ENPP3 | 100 | 81.829 | ENSBTAG00000020196 | ENPP3 | 100 | 81.829 | Bos_taurus |
| ENSG00000154269 | ENPP3 | 100 | 94.171 | ENSCJAG00000003044 | ENPP3 | 100 | 94.171 | Callithrix_jacchus |
| ENSG00000154269 | ENPP3 | 100 | 84.949 | ENSCAFG00000000367 | ENPP3 | 100 | 84.949 | Canis_familiaris |
| ENSG00000154269 | ENPP3 | 98 | 82.305 | ENSCAFG00020017451 | ENPP3 | 99 | 82.189 | Canis_lupus_dingo |
| ENSG00000154269 | ENPP3 | 100 | 81.600 | ENSCHIG00000000508 | ENPP3 | 100 | 81.600 | Capra_hircus |
| ENSG00000154269 | ENPP3 | 100 | 86.057 | ENSTSYG00000006067 | ENPP3 | 100 | 86.057 | Carlito_syrichta |
| ENSG00000154269 | ENPP3 | 95 | 64.881 | ENSCAPG00000015290 | ENPP3 | 100 | 64.881 | Cavia_aperea |
| ENSG00000154269 | ENPP3 | 95 | 68.585 | ENSCPOG00000038769 | ENPP3 | 100 | 68.705 | Cavia_porcellus |
| ENSG00000154269 | ENPP3 | 100 | 94.312 | ENSCCAG00000036027 | ENPP3 | 100 | 94.312 | Cebus_capucinus |
| ENSG00000154269 | ENPP3 | 100 | 94.749 | ENSCATG00000044442 | ENPP3 | 100 | 94.749 | Cercocebus_atys |
| ENSG00000154269 | ENPP3 | 100 | 78.107 | ENSCLAG00000009551 | ENPP3 | 99 | 78.107 | Chinchilla_lanigera |
| ENSG00000154269 | ENPP3 | 100 | 96.686 | ENSCSAG00000016657 | ENPP3 | 93 | 96.686 | Chlorocebus_sabaeus |
| ENSG00000154269 | ENPP3 | 99 | 68.270 | ENSCPBG00000017819 | ENPP3 | 98 | 68.270 | Chrysemys_picta_bellii |
| ENSG00000154269 | ENPP3 | 100 | 95.314 | ENSCANG00000034260 | ENPP3 | 100 | 95.314 | Colobus_angolensis_palliatus |
| ENSG00000154269 | ENPP3 | 100 | 81.849 | ENSCGRG00001001049 | Enpp3 | 100 | 81.849 | Cricetulus_griseus_chok1gshd |
| ENSG00000154269 | ENPP3 | 100 | 82.098 | ENSDNOG00000014248 | ENPP3 | 100 | 82.098 | Dasypus_novemcinctus |
| ENSG00000154269 | ENPP3 | 100 | 78.977 | ENSDORG00000008720 | Enpp3 | 100 | 78.977 | Dipodomys_ordii |
| ENSG00000154269 | ENPP3 | 76 | 79.455 | ENSETEG00000007816 | - | 76 | 79.455 | Echinops_telfairi |
| ENSG00000154269 | ENPP3 | 100 | 83.886 | ENSECAG00000007018 | ENPP3 | 100 | 83.886 | Equus_caballus |
| ENSG00000154269 | ENPP3 | 100 | 82.286 | ENSECAG00000012424 | - | 100 | 82.286 | Equus_caballus |
| ENSG00000154269 | ENPP3 | 70 | 85.268 | ENSEEUG00000000695 | - | 72 | 85.268 | Erinaceus_europaeus |
| ENSG00000154269 | ENPP3 | 100 | 84.571 | ENSFCAG00000014130 | ENPP3 | 100 | 84.571 | Felis_catus |
| ENSG00000154269 | ENPP3 | 83 | 62.364 | ENSFALG00000011732 | ENPP3 | 99 | 62.364 | Ficedula_albicollis |
| ENSG00000154269 | ENPP3 | 100 | 75.000 | ENSFDAG00000011940 | ENPP3 | 100 | 75.000 | Fukomys_damarensis |
| ENSG00000154269 | ENPP3 | 99 | 66.551 | ENSGALG00000037587 | ENPP3 | 97 | 66.319 | Gallus_gallus |
| ENSG00000154269 | ENPP3 | 97 | 66.470 | ENSGAGG00000022901 | ENPP3 | 99 | 70.325 | Gopherus_agassizii |
| ENSG00000154269 | ENPP3 | 100 | 78.539 | ENSHGLG00000013966 | ENPP3 | 100 | 78.539 | Heterocephalus_glaber_female |
| ENSG00000154269 | ENPP3 | 100 | 78.539 | ENSHGLG00100018706 | ENPP3 | 100 | 78.539 | Heterocephalus_glaber_male |
| ENSG00000154269 | ENPP3 | 100 | 81.829 | ENSJJAG00000019911 | Enpp3 | 100 | 81.829 | Jaculus_jaculus |
| ENSG00000154269 | ENPP3 | 100 | 83.771 | ENSLAFG00000003881 | ENPP3 | 100 | 83.771 | Loxodonta_africana |
| ENSG00000154269 | ENPP3 | 100 | 97.029 | ENSMFAG00000040950 | ENPP3 | 100 | 97.029 | Macaca_fascicularis |
| ENSG00000154269 | ENPP3 | 100 | 97.143 | ENSMMUG00000017549 | ENPP3 | 100 | 97.143 | Macaca_mulatta |
| ENSG00000154269 | ENPP3 | 100 | 96.800 | ENSMNEG00000034089 | ENPP3 | 100 | 96.800 | Macaca_nemestrina |
| ENSG00000154269 | ENPP3 | 100 | 96.914 | ENSMLEG00000028918 | ENPP3 | 100 | 96.914 | Mandrillus_leucophaeus |
| ENSG00000154269 | ENPP3 | 99 | 65.358 | ENSMGAG00000013156 | ENPP3 | 99 | 66.166 | Meleagris_gallopavo |
| ENSG00000154269 | ENPP3 | 100 | 74.315 | ENSMAUG00000001245 | Enpp3 | 100 | 74.315 | Mesocricetus_auratus |
| ENSG00000154269 | ENPP3 | 100 | 88.000 | ENSMICG00000002944 | ENPP3 | 100 | 88.000 | Microcebus_murinus |
| ENSG00000154269 | ENPP3 | 100 | 76.444 | ENSMOCG00000003824 | Enpp3 | 100 | 76.444 | Microtus_ochrogaster |
| ENSG00000154269 | ENPP3 | 98 | 70.788 | ENSMODG00000017569 | ENPP3 | 100 | 69.575 | Monodelphis_domestica |
| ENSG00000154269 | ENPP3 | 100 | 80.251 | MGP_CAROLIEiJ_G0015190 | Enpp3 | 100 | 80.251 | Mus_caroli |
| ENSG00000154269 | ENPP3 | 100 | 80.708 | ENSMUSG00000019989 | Enpp3 | 100 | 80.708 | Mus_musculus |
| ENSG00000154269 | ENPP3 | 98 | 76.117 | MGP_PahariEiJ_G0023812 | Enpp3 | 100 | 73.378 | Mus_pahari |
| ENSG00000154269 | ENPP3 | 100 | 80.708 | MGP_SPRETEiJ_G0015999 | Enpp3 | 96 | 84.713 | Mus_spretus |
| ENSG00000154269 | ENPP3 | 100 | 84.055 | ENSMPUG00000014671 | ENPP3 | 100 | 84.055 | Mustela_putorius_furo |
| ENSG00000154269 | ENPP3 | 100 | 80.914 | ENSMLUG00000016827 | ENPP3 | 100 | 80.914 | Myotis_lucifugus |
| ENSG00000154269 | ENPP3 | 99 | 82.503 | ENSNGAG00000015161 | Enpp3 | 99 | 82.503 | Nannospalax_galili |
| ENSG00000154269 | ENPP3 | 100 | 97.029 | ENSNLEG00000014455 | ENPP3 | 100 | 97.029 | Nomascus_leucogenys |
| ENSG00000154269 | ENPP3 | 99 | 70.183 | ENSOPRG00000002617 | ENPP3 | 100 | 70.183 | Ochotona_princeps |
| ENSG00000154269 | ENPP3 | 100 | 82.743 | ENSOCUG00000014432 | ENPP3 | 100 | 82.743 | Oryctolagus_cuniculus |
| ENSG00000154269 | ENPP3 | 100 | 81.486 | ENSOARG00000014082 | ENPP3 | 97 | 81.486 | Ovis_aries |
| ENSG00000154269 | ENPP3 | 100 | 99.200 | ENSPPAG00000003107 | ENPP3 | 100 | 99.200 | Pan_paniscus |
| ENSG00000154269 | ENPP3 | 100 | 84.800 | ENSPPRG00000023946 | ENPP3 | 100 | 84.800 | Panthera_pardus |
| ENSG00000154269 | ENPP3 | 100 | 84.800 | ENSPTIG00000012163 | ENPP3 | 100 | 84.800 | Panthera_tigris_altaica |
| ENSG00000154269 | ENPP3 | 100 | 99.314 | ENSPTRG00000023128 | ENPP3 | 100 | 99.314 | Pan_troglodytes |
| ENSG00000154269 | ENPP3 | 100 | 96.914 | ENSPANG00000006092 | ENPP3 | 93 | 96.914 | Papio_anubis |
| ENSG00000154269 | ENPP3 | 82 | 70.694 | ENSPSIG00000006898 | ENPP3 | 98 | 70.694 | Pelodiscus_sinensis |
| ENSG00000154269 | ENPP3 | 100 | 80.844 | ENSPEMG00000022836 | Enpp3 | 100 | 80.844 | Peromyscus_maniculatus_bairdii |
| ENSG00000154269 | ENPP3 | 97 | 52.693 | ENSPMAG00000002458 | ENPP3 | 100 | 52.693 | Petromyzon_marinus |
| ENSG00000154269 | ENPP3 | 98 | 73.457 | ENSPCIG00000008801 | ENPP3 | 96 | 73.138 | Phascolarctos_cinereus |
| ENSG00000154269 | ENPP3 | 100 | 83.543 | ENSPPYG00000017009 | ENPP3 | 100 | 83.543 | Pongo_abelii |
| ENSG00000154269 | ENPP3 | 100 | 74.514 | ENSPCOG00000017134 | ENPP3 | 100 | 74.514 | Propithecus_coquereli |
| ENSG00000154269 | ENPP3 | 100 | 74.800 | ENSPVAG00000013515 | ENPP3 | 100 | 74.800 | Pteropus_vampyrus |
| ENSG00000154269 | ENPP3 | 100 | 81.393 | ENSRNOG00000013791 | Enpp3 | 100 | 81.393 | Rattus_norvegicus |
| ENSG00000154269 | ENPP3 | 100 | 96.114 | ENSRBIG00000030207 | ENPP3 | 99 | 96.114 | Rhinopithecus_bieti |
| ENSG00000154269 | ENPP3 | 100 | 96.571 | ENSRROG00000035272 | ENPP3 | 100 | 96.571 | Rhinopithecus_roxellana |
| ENSG00000154269 | ENPP3 | 100 | 94.057 | ENSSBOG00000034636 | ENPP3 | 100 | 94.057 | Saimiri_boliviensis_boliviensis |
| ENSG00000154269 | ENPP3 | 99 | 72.426 | ENSSHAG00000008503 | ENPP3 | 99 | 72.426 | Sarcophilus_harrisii |
| ENSG00000154269 | ENPP3 | 96 | 62.500 | ENSSPUG00000007266 | ENPP3 | 97 | 64.295 | Sphenodon_punctatus |
| ENSG00000154269 | ENPP3 | 100 | 83.314 | ENSSSCG00000025992 | ENPP3 | 100 | 83.314 | Sus_scrofa |
| ENSG00000154269 | ENPP3 | 99 | 66.246 | ENSTGUG00000011640 | ENPP3 | 99 | 66.016 | Taeniopygia_guttata |
| ENSG00000154269 | ENPP3 | 100 | 75.228 | ENSTBEG00000009399 | ENPP3 | 100 | 75.228 | Tupaia_belangeri |
| ENSG00000154269 | ENPP3 | 100 | 82.286 | ENSTTRG00000011371 | ENPP3 | 100 | 82.286 | Tursiops_truncatus |
| ENSG00000154269 | ENPP3 | 100 | 81.257 | ENSUAMG00000022974 | ENPP3 | 100 | 82.195 | Ursus_americanus |
| ENSG00000154269 | ENPP3 | 100 | 84.114 | ENSUMAG00000024546 | ENPP3 | 100 | 84.114 | Ursus_maritimus |
| ENSG00000154269 | ENPP3 | 100 | 80.571 | ENSVPAG00000005657 | ENPP3 | 100 | 80.571 | Vicugna_pacos |
| ENSG00000154269 | ENPP3 | 97 | 82.629 | ENSVVUG00000009079 | - | 99 | 84.727 | Vulpes_vulpes |
| ENSG00000154269 | ENPP3 | 97 | 61.702 | ENSXETG00000008244 | enpp3 | 99 | 61.910 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002276 | basophil activation involved in immune response | - | ISS | Process |
| GO:0003676 | nucleic acid binding | - | IEA | Function |
| GO:0004528 | phosphodiesterase I activity | 11342463. | IMP | Function |
| GO:0005044 | scavenger receptor activity | - | IEA | Function |
| GO:0005509 | calcium ion binding | 29717535. | IDA | Function |
| GO:0006220 | pyrimidine nucleotide metabolic process | 11342463. | IMP | Process |
| GO:0006796 | phosphate-containing compound metabolic process | 10513816. | IDA | Process |
| GO:0006898 | receptor-mediated endocytosis | - | IEA | Process |
| GO:0008270 | zinc ion binding | 29717535. | IDA | Function |
| GO:0009143 | nucleoside triphosphate catabolic process | 21873635. | IBA | Process |
| GO:0009143 | nucleoside triphosphate catabolic process | 10513816. | IDA | Process |
| GO:0009897 | external side of plasma membrane | 11342463. | IDA | Component |
| GO:0016021 | integral component of membrane | - | IEA | Component |
| GO:0016324 | apical plasma membrane | - | IEA | Component |
| GO:0030247 | polysaccharide binding | - | IEA | Function |
| GO:0030505 | inorganic diphosphate transport | 10513816. | IDA | Process |
| GO:0033007 | negative regulation of mast cell activation involved in immune response | - | ISS | Process |
| GO:0035529 | NADH pyrophosphatase activity | - | IEA | Function |
| GO:0046034 | ATP metabolic process | 29717535. | IDA | Process |
| GO:0047429 | nucleoside-triphosphate diphosphatase activity | 21873635. | IBA | Function |
| GO:0047429 | nucleoside-triphosphate diphosphatase activity | 10513816.29717535. | IDA | Function |
| GO:0048471 | perinuclear region of cytoplasm | 10513816. | IDA | Component |
| GO:0050728 | negative regulation of inflammatory response | - | ISS | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708. | HDA | Component |
| GO:0070667 | negative regulation of mast cell proliferation | - | ISS | Process |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | - | IEA | Process |