
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| KIRP | |||
| LAML | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| MESO | ||
| HNSC | ||
| COAD | ||
| BLCA | ||
| CESC | ||
| READ | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000409364 | RRM_1 | PF00076.22 | 8.3e-08 | 1 | 1 |
| ENSP00000384403 | RRM_1 | PF00076.22 | 1.1e-07 | 1 | 1 |
| ENSP00000353638 | RRM_1 | PF00076.22 | 1.1e-07 | 1 | 1 |
| ENSP00000377855 | RRM_1 | PF00076.22 | 1.2e-07 | 1 | 1 |
| ENSP00000312649 | RRM_1 | PF00076.22 | 1.2e-07 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000360453 | PPARGC1B-202 | 3004 | XM_011537555 | ENSP00000353638 | 984 (aa) | XP_011535857 | Q86YN6 |
| ENST00000461780 | PPARGC1B-206 | 566 | - | - | - (aa) | - | - |
| ENST00000403750 | PPARGC1B-204 | 3146 | - | ENSP00000384403 | 959 (aa) | - | Q86YN6 |
| ENST00000394320 | PPARGC1B-203 | 4803 | XM_011537553 | ENSP00000377855 | 1017 (aa) | XP_011535855 | Q86YN6 |
| ENST00000309241 | PPARGC1B-201 | 10568 | XM_005268372 | ENSP00000312649 | 1023 (aa) | XP_005268429 | Q86YN6 |
| ENST00000492495 | PPARGC1B-207 | 572 | - | - | - (aa) | - | - |
| ENST00000434684 | PPARGC1B-205 | 2457 | XM_011537556 | ENSP00000409364 | 748 (aa) | XP_011535858 | H0Y713 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04931 | Insulin resistance | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000155846 | Triglycerides | 2.09759943155418E-6 | 17903299 |
| ENSG00000155846 | Lymphocyte Count | 9.243e-006 | - |
| ENSG00000155846 | Lymphocyte Count | 8.544e-005 | - |
| ENSG00000155846 | Lymphocyte Count | 9.463e-007 | - |
| ENSG00000155846 | Neutrophils | 8.618e-005 | - |
| ENSG00000155846 | Neutrophils | 1.748e-005 | - |
| ENSG00000155846 | Platelet Function Tests | 1.5809800E-005 | - |
| ENSG00000155846 | Platelet Function Tests | 1.8800000E-006 | - |
| ENSG00000155846 | Platelet Function Tests | 3.3400000E-006 | - |
| ENSG00000155846 | Platelet Function Tests | 2.3637900E-005 | - |
| ENSG00000155846 | Platelet Function Tests | 1.9419200E-005 | - |
| ENSG00000155846 | Fenofibrate | 3E-7 | 22890011 |
| ENSG00000155846 | Suntan | 4E-6 | 19340012 |
| ENSG00000155846 | Estradiol | 5E-6 | 23251661 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000155846 | rs251464 | 5 | 149816671 | C | Low tan response | 29739929 | [0.011-0.109] unit decrease | 0.06 | EFO_0004279 |
| ENSG00000155846 | rs9285640 | 5 | 149762504 | G | Response to fenofibrate | 22890011 | [18.62-41.28] unit decrease | 29.95 | GO_1901557 |
| ENSG00000155846 | rs32579 | 5 | 149831285 | A | Tanning | 19340012 | [0.07-0.15] tanning ability score decrease | 0.11 | EFO_0004279 |
| ENSG00000155846 | rs32576 | 5 | 149833893 | G | Obesity-related traits | 23251661 | [NR] pg/mL increase | 0.03 | EFO_0004697 |
| ENSG00000155846 | rs251464 | 5 | 149816671 | ? | Vitiligo | 27723757 | NR | 1.155 | EFO_0004208 |
| ENSG00000155846 | rs251468 | 5 | 149814922 | C | Age spots | 29895819 | [1.2-1.39] | 1.2987013 | EFO_0007850 |
| ENSG00000155846 | rs251468 | 5 | 149814922 | C | Freckles | 29895819 | [1.33-1.56] | 1.4492754 | EFO_0003963 |
| ENSG00000155846 | rs251464 | 5 | 149816671 | ? | Nevus count or cutaneous melanoma | 30429480 | EFO_0004632|EFO_0000389 | ||
| ENSG00000155846 | rs251464 | 5 | 149816671 | ? | Nevus count | 30429480 | EFO_0004632 | ||
| ENSG00000155846 | rs251466 | 5 | 149816040 | ? | Sunburns | 30595370 | EFO_0003958 | ||
| ENSG00000155846 | rs13174179 | 5 | 149771108 | ? | Lung function (FEV1/FVC) | 30595370 | EFO_0004713 |
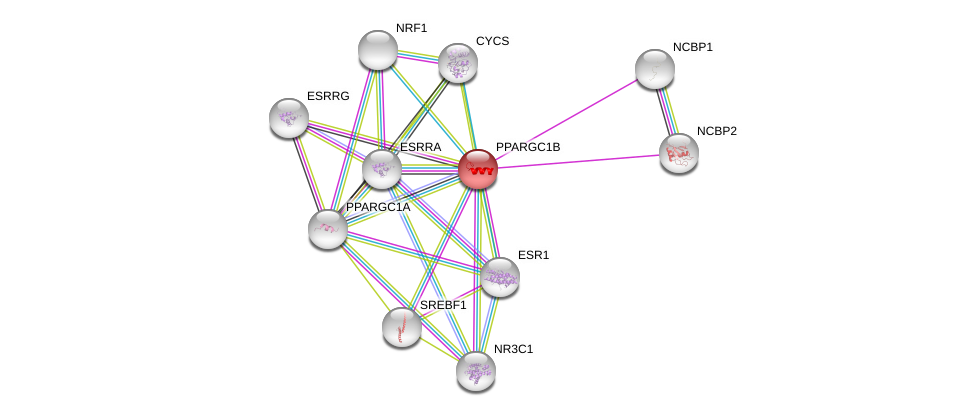
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSG00000109819 | PPARGC1A | 55 | 51.908 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000155846 | PPARGC1B | 50 | 56.489 | ENSAPOG00000018632 | ppargc1a | 68 | 56.489 | Acanthochromis_polyacanthus |
| ENSG00000155846 | PPARGC1B | 100 | 84.705 | ENSAMEG00000005287 | PPARGC1B | 100 | 84.705 | Ailuropoda_melanoleuca |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSAMEG00000010421 | PPARGC1A | 55 | 51.908 | Ailuropoda_melanoleuca |
| ENSG00000155846 | PPARGC1B | 50 | 56.489 | ENSAOCG00000006754 | ppargc1a | 68 | 56.489 | Amphiprion_ocellaris |
| ENSG00000155846 | PPARGC1B | 50 | 56.489 | ENSAPEG00000022574 | ppargc1a | 67 | 56.489 | Amphiprion_percula |
| ENSG00000155846 | PPARGC1B | 99 | 49.521 | ENSAPLG00000009977 | PPARGC1B | 99 | 48.954 | Anas_platyrhynchos |
| ENSG00000155846 | PPARGC1B | 100 | 48.155 | ENSACAG00000015737 | PPARGC1B | 99 | 47.527 | Anolis_carolinensis |
| ENSG00000155846 | PPARGC1B | 52 | 52.672 | ENSACAG00000000741 | PPARGC1A | 56 | 52.672 | Anolis_carolinensis |
| ENSG00000155846 | PPARGC1B | 100 | 93.652 | ENSANAG00000021830 | PPARGC1B | 100 | 93.750 | Aotus_nancymaae |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSANAG00000035930 | PPARGC1A | 57 | 51.908 | Aotus_nancymaae |
| ENSG00000155846 | PPARGC1B | 51 | 54.198 | ENSAMXG00000002461 | ppargc1a | 65 | 54.198 | Astyanax_mexicanus |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSBTAG00000017024 | PPARGC1A | 56 | 51.145 | Bos_taurus |
| ENSG00000155846 | PPARGC1B | 100 | 82.285 | ENSBTAG00000012943 | PPARGC1B | 100 | 82.478 | Bos_taurus |
| ENSG00000155846 | PPARGC1B | 100 | 92.780 | ENSCJAG00000021113 | PPARGC1B | 100 | 91.477 | Callithrix_jacchus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSCJAG00000013024 | PPARGC1A | 53 | 51.908 | Callithrix_jacchus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSCAFG00000016519 | PPARGC1A | 55 | 51.908 | Canis_familiaris |
| ENSG00000155846 | PPARGC1B | 100 | 85.010 | ENSCAFG00000018275 | PPARGC1B | 100 | 84.816 | Canis_familiaris |
| ENSG00000155846 | PPARGC1B | 100 | 84.824 | ENSCAFG00020023524 | PPARGC1B | 100 | 84.623 | Canis_lupus_dingo |
| ENSG00000155846 | PPARGC1B | 100 | 81.801 | ENSCHIG00000004409 | PPARGC1B | 100 | 81.994 | Capra_hircus |
| ENSG00000155846 | PPARGC1B | 56 | 52.308 | ENSCHIG00000015991 | PPARGC1A | 56 | 52.308 | Capra_hircus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSTSYG00000010439 | PPARGC1A | 55 | 51.908 | Carlito_syrichta |
| ENSG00000155846 | PPARGC1B | 99 | 77.894 | ENSTSYG00000034354 | PPARGC1B | 100 | 78.519 | Carlito_syrichta |
| ENSG00000155846 | PPARGC1B | 99 | 79.607 | ENSCAPG00000008859 | PPARGC1B | 99 | 79.628 | Cavia_aperea |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSCPOG00000001518 | PPARGC1A | 55 | 51.908 | Cavia_porcellus |
| ENSG00000155846 | PPARGC1B | 100 | 79.517 | ENSCPOG00000001958 | PPARGC1B | 100 | 79.556 | Cavia_porcellus |
| ENSG00000155846 | PPARGC1B | 100 | 93.848 | ENSCCAG00000026909 | PPARGC1B | 100 | 93.555 | Cebus_capucinus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSCCAG00000024681 | PPARGC1A | 57 | 51.908 | Cebus_capucinus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSCATG00000032575 | PPARGC1A | 58 | 51.908 | Cercocebus_atys |
| ENSG00000155846 | PPARGC1B | 100 | 96.240 | ENSCATG00000030402 | PPARGC1B | 100 | 96.240 | Cercocebus_atys |
| ENSG00000155846 | PPARGC1B | 100 | 79.074 | ENSCLAG00000015452 | PPARGC1B | 100 | 79.267 | Chinchilla_lanigera |
| ENSG00000155846 | PPARGC1B | 52 | 48.052 | ENSCLAG00000002058 | PPARGC1A | 58 | 51.042 | Chinchilla_lanigera |
| ENSG00000155846 | PPARGC1B | 98 | 96.092 | ENSCSAG00000011792 | PPARGC1B | 100 | 96.092 | Chlorocebus_sabaeus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSCSAG00000016575 | PPARGC1A | 55 | 51.908 | Chlorocebus_sabaeus |
| ENSG00000155846 | PPARGC1B | 63 | 88.095 | ENSCHOG00000005517 | - | 68 | 88.095 | Choloepus_hoffmanni |
| ENSG00000155846 | PPARGC1B | 99 | 50.337 | ENSCPBG00000007891 | PPARGC1B | 99 | 49.529 | Chrysemys_picta_bellii |
| ENSG00000155846 | PPARGC1B | 100 | 95.122 | ENSCANG00000037487 | PPARGC1B | 100 | 95.220 | Colobus_angolensis_palliatus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSCANG00000033426 | PPARGC1A | 58 | 51.908 | Colobus_angolensis_palliatus |
| ENSG00000155846 | PPARGC1B | 50 | 50.382 | ENSCGRG00001015534 | Ppargc1a | 55 | 50.382 | Cricetulus_griseus_chok1gshd |
| ENSG00000155846 | PPARGC1B | 97 | 77.978 | ENSCGRG00001017949 | Ppargc1b | 96 | 77.978 | Cricetulus_griseus_chok1gshd |
| ENSG00000155846 | PPARGC1B | 98 | 77.323 | ENSCGRG00000008387 | Ppargc1b | 99 | 77.323 | Cricetulus_griseus_crigri |
| ENSG00000155846 | PPARGC1B | 50 | 50.382 | ENSCGRG00000000200 | Ppargc1a | 55 | 50.382 | Cricetulus_griseus_crigri |
| ENSG00000155846 | PPARGC1B | 50 | 54.198 | ENSCSEG00000006884 | ppargc1a | 67 | 54.198 | Cynoglossus_semilaevis |
| ENSG00000155846 | PPARGC1B | 61 | 42.063 | ENSDARG00000101569 | ppargc1b | 79 | 43.651 | Danio_rerio |
| ENSG00000155846 | PPARGC1B | 51 | 54.198 | ENSDARG00000067829 | ppargc1a | 66 | 54.198 | Danio_rerio |
| ENSG00000155846 | PPARGC1B | 95 | 85.627 | ENSDNOG00000005985 | PPARGC1B | 99 | 85.321 | Dasypus_novemcinctus |
| ENSG00000155846 | PPARGC1B | 100 | 79.192 | ENSDORG00000012605 | Ppargc1b | 100 | 79.394 | Dipodomys_ordii |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSDORG00000016276 | Ppargc1a | 58 | 51.042 | Dipodomys_ordii |
| ENSG00000155846 | PPARGC1B | 100 | 84.100 | ENSEASG00005002826 | PPARGC1B | 100 | 83.600 | Equus_asinus_asinus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSEASG00005007984 | PPARGC1A | 55 | 51.908 | Equus_asinus_asinus |
| ENSG00000155846 | PPARGC1B | 100 | 83.622 | ENSECAG00000019769 | PPARGC1B | 83 | 83.815 | Equus_caballus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSECAG00000009164 | PPARGC1A | 55 | 51.908 | Equus_caballus |
| ENSG00000155846 | PPARGC1B | 51 | 48.739 | ENSEEUG00000009219 | PPARGC1A | 57 | 52.672 | Erinaceus_europaeus |
| ENSG00000155846 | PPARGC1B | 80 | 74.348 | ENSEEUG00000014903 | PPARGC1B | 86 | 76.087 | Erinaceus_europaeus |
| ENSG00000155846 | PPARGC1B | 75 | 55.000 | ENSELUG00000014929 | ppargc1b | 74 | 44.094 | Esox_lucius |
| ENSG00000155846 | PPARGC1B | 50 | 49.550 | ENSFCAG00000000043 | PPARGC1A | 63 | 51.908 | Felis_catus |
| ENSG00000155846 | PPARGC1B | 99 | 47.843 | ENSFALG00000008858 | PPARGC1B | 99 | 50.489 | Ficedula_albicollis |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSFDAG00000005659 | PPARGC1A | 55 | 51.908 | Fukomys_damarensis |
| ENSG00000155846 | PPARGC1B | 97 | 78.734 | ENSFDAG00000008719 | PPARGC1B | 99 | 78.932 | Fukomys_damarensis |
| ENSG00000155846 | PPARGC1B | 51 | 53.435 | ENSGALG00000042851 | PPARGC1A | 57 | 53.435 | Gallus_gallus |
| ENSG00000155846 | PPARGC1B | 91 | 74.026 | ENSGAGG00000012870 | PPARGC1B | 98 | 72.840 | Gopherus_agassizii |
| ENSG00000155846 | PPARGC1B | 58 | 49.580 | ENSGAGG00000007043 | PPARGC1A | 55 | 53.435 | Gopherus_agassizii |
| ENSG00000155846 | PPARGC1B | 100 | 98.540 | ENSGGOG00000009505 | PPARGC1B | 100 | 98.596 | Gorilla_gorilla |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSGGOG00000009810 | PPARGC1A | 56 | 51.908 | Gorilla_gorilla |
| ENSG00000155846 | PPARGC1B | 100 | 80.485 | ENSHGLG00000005967 | PPARGC1B | 100 | 80.874 | Heterocephalus_glaber_female |
| ENSG00000155846 | PPARGC1B | 50 | 52.672 | ENSHGLG00000007022 | PPARGC1A | 56 | 52.672 | Heterocephalus_glaber_female |
| ENSG00000155846 | PPARGC1B | 50 | 52.672 | ENSHGLG00100000133 | PPARGC1A | 55 | 52.672 | Heterocephalus_glaber_male |
| ENSG00000155846 | PPARGC1B | 100 | 79.474 | ENSHGLG00100010588 | PPARGC1B | 100 | 79.879 | Heterocephalus_glaber_male |
| ENSG00000155846 | PPARGC1B | 100 | 83.930 | ENSSTOG00000024758 | PPARGC1B | 100 | 84.511 | Ictidomys_tridecemlineatus |
| ENSG00000155846 | PPARGC1B | 51 | 52.672 | ENSSTOG00000012522 | PPARGC1A | 55 | 52.672 | Ictidomys_tridecemlineatus |
| ENSG00000155846 | PPARGC1B | 100 | 78.613 | ENSJJAG00000018633 | Ppargc1b | 100 | 78.870 | Jaculus_jaculus |
| ENSG00000155846 | PPARGC1B | 99 | 36.266 | ENSLACG00000011232 | PPARGC1B | 100 | 36.616 | Latimeria_chalumnae |
| ENSG00000155846 | PPARGC1B | 52 | 58.015 | ENSLACG00000014054 | PPARGC1A | 58 | 58.015 | Latimeria_chalumnae |
| ENSG00000155846 | PPARGC1B | 69 | 46.667 | ENSLOCG00000012195 | ppargc1b | 89 | 46.667 | Lepisosteus_oculatus |
| ENSG00000155846 | PPARGC1B | 52 | 56.489 | ENSLOCG00000012350 | ppargc1a | 68 | 56.489 | Lepisosteus_oculatus |
| ENSG00000155846 | PPARGC1B | 98 | 81.405 | ENSLAFG00000012339 | PPARGC1B | 99 | 80.601 | Loxodonta_africana |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSMFAG00000000534 | PPARGC1A | 58 | 51.908 | Macaca_fascicularis |
| ENSG00000155846 | PPARGC1B | 100 | 96.188 | ENSMFAG00000042269 | PPARGC1B | 100 | 96.188 | Macaca_fascicularis |
| ENSG00000155846 | PPARGC1B | 100 | 96.188 | ENSMMUG00000004749 | PPARGC1B | 100 | 96.188 | Macaca_mulatta |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSMMUG00000001844 | PPARGC1A | 55 | 51.908 | Macaca_mulatta |
| ENSG00000155846 | PPARGC1B | 100 | 96.090 | ENSMNEG00000034121 | PPARGC1B | 100 | 96.090 | Macaca_nemestrina |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSMNEG00000028682 | PPARGC1A | 58 | 51.908 | Macaca_nemestrina |
| ENSG00000155846 | PPARGC1B | 97 | 96.088 | ENSMLEG00000040973 | PPARGC1B | 100 | 96.088 | Mandrillus_leucophaeus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSMLEG00000034233 | PPARGC1A | 58 | 51.908 | Mandrillus_leucophaeus |
| ENSG00000155846 | PPARGC1B | 51 | 54.198 | ENSMGAG00000012485 | PPARGC1A | 56 | 54.198 | Meleagris_gallopavo |
| ENSG00000155846 | PPARGC1B | 84 | 70.161 | ENSMGAG00000003134 | PPARGC1B | 81 | 70.866 | Meleagris_gallopavo |
| ENSG00000155846 | PPARGC1B | 100 | 77.388 | ENSMAUG00000014191 | Ppargc1b | 100 | 77.290 | Mesocricetus_auratus |
| ENSG00000155846 | PPARGC1B | 50 | 50.370 | ENSMAUG00000011811 | Ppargc1a | 56 | 50.370 | Mesocricetus_auratus |
| ENSG00000155846 | PPARGC1B | 100 | 85.352 | ENSMICG00000002702 | PPARGC1B | 100 | 85.352 | Microcebus_murinus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSMICG00000015390 | PPARGC1A | 59 | 51.908 | Microcebus_murinus |
| ENSG00000155846 | PPARGC1B | 100 | 77.810 | ENSMOCG00000014885 | Ppargc1b | 100 | 77.713 | Microtus_ochrogaster |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSMOCG00000008765 | Ppargc1a | 55 | 51.145 | Microtus_ochrogaster |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSMODG00000000549 | PPARGC1A | 55 | 52.083 | Monodelphis_domestica |
| ENSG00000155846 | PPARGC1B | 98 | 58.006 | ENSMODG00000009891 | PPARGC1B | 99 | 57.674 | Monodelphis_domestica |
| ENSG00000155846 | PPARGC1B | 100 | 78.328 | MGP_CAROLIEiJ_G0022323 | Ppargc1b | 100 | 78.134 | Mus_caroli |
| ENSG00000155846 | PPARGC1B | 50 | 50.382 | MGP_CAROLIEiJ_G0027213 | Ppargc1a | 55 | 51.087 | Mus_caroli |
| ENSG00000155846 | PPARGC1B | 100 | 78.384 | ENSMUSG00000033871 | Ppargc1b | 100 | 78.189 | Mus_musculus |
| ENSG00000155846 | PPARGC1B | 50 | 50.382 | ENSMUSG00000029167 | Ppargc1a | 55 | 51.087 | Mus_musculus |
| ENSG00000155846 | PPARGC1B | 52 | 47.403 | MGP_PahariEiJ_G0016995 | Ppargc1a | 57 | 51.087 | Mus_pahari |
| ENSG00000155846 | PPARGC1B | 100 | 76.544 | MGP_PahariEiJ_G0019053 | Ppargc1b | 100 | 76.641 | Mus_pahari |
| ENSG00000155846 | PPARGC1B | 50 | 50.382 | MGP_SPRETEiJ_G0028208 | Ppargc1a | 55 | 51.087 | Mus_spretus |
| ENSG00000155846 | PPARGC1B | 100 | 78.092 | MGP_SPRETEiJ_G0023235 | Ppargc1b | 100 | 77.897 | Mus_spretus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSMPUG00000016736 | PPARGC1A | 55 | 51.908 | Mustela_putorius_furo |
| ENSG00000155846 | PPARGC1B | 100 | 84.608 | ENSMPUG00000014245 | PPARGC1B | 100 | 84.608 | Mustela_putorius_furo |
| ENSG00000155846 | PPARGC1B | 92 | 77.406 | ENSMLUG00000013169 | PPARGC1B | 100 | 77.301 | Myotis_lucifugus |
| ENSG00000155846 | PPARGC1B | 51 | 48.739 | ENSMLUG00000016307 | PPARGC1A | 55 | 51.908 | Myotis_lucifugus |
| ENSG00000155846 | PPARGC1B | 99 | 78.732 | ENSNGAG00000016998 | Ppargc1b | 99 | 78.439 | Nannospalax_galili |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSNGAG00000015090 | Ppargc1a | 55 | 51.145 | Nannospalax_galili |
| ENSG00000155846 | PPARGC1B | 100 | 94.824 | ENSNLEG00000010193 | PPARGC1B | 100 | 94.824 | Nomascus_leucogenys |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSNLEG00000016760 | PPARGC1A | 57 | 51.908 | Nomascus_leucogenys |
| ENSG00000155846 | PPARGC1B | 97 | 75.651 | ENSOPRG00000010017 | PPARGC1B | 99 | 75.651 | Ochotona_princeps |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSOPRG00000012899 | PPARGC1A | 55 | 51.145 | Ochotona_princeps |
| ENSG00000155846 | PPARGC1B | 100 | 80.000 | ENSODEG00000004901 | PPARGC1B | 100 | 80.303 | Octodon_degus |
| ENSG00000155846 | PPARGC1B | 50 | 52.672 | ENSOANG00000006680 | PPARGC1A | 55 | 52.672 | Ornithorhynchus_anatinus |
| ENSG00000155846 | PPARGC1B | 99 | 54.618 | ENSOANG00000009368 | PPARGC1B | 99 | 55.120 | Ornithorhynchus_anatinus |
| ENSG00000155846 | PPARGC1B | 98 | 78.381 | ENSOCUG00000013833 | PPARGC1B | 100 | 78.381 | Oryctolagus_cuniculus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSOCUG00000014668 | PPARGC1A | 55 | 51.908 | Oryctolagus_cuniculus |
| ENSG00000155846 | PPARGC1B | 98 | 80.915 | ENSOGAG00000015983 | PPARGC1B | 99 | 79.825 | Otolemur_garnettii |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSOGAG00000014559 | PPARGC1A | 55 | 51.145 | Otolemur_garnettii |
| ENSG00000155846 | PPARGC1B | 98 | 80.000 | ENSOARG00000005658 | PPARGC1B | 98 | 80.000 | Ovis_aries |
| ENSG00000155846 | PPARGC1B | 50 | 52.308 | ENSOARG00000005581 | PPARGC1A | 54 | 52.308 | Ovis_aries |
| ENSG00000155846 | PPARGC1B | 100 | 99.166 | ENSPPAG00000035226 | PPARGC1B | 100 | 99.198 | Pan_paniscus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSPPAG00000034694 | PPARGC1A | 60 | 51.908 | Pan_paniscus |
| ENSG00000155846 | PPARGC1B | 100 | 86.227 | ENSPPRG00000004608 | PPARGC1B | 100 | 86.421 | Panthera_pardus |
| ENSG00000155846 | PPARGC1B | 50 | 49.550 | ENSPPRG00000001711 | PPARGC1A | 63 | 51.908 | Panthera_pardus |
| ENSG00000155846 | PPARGC1B | 99 | 85.227 | ENSPTIG00000007391 | PPARGC1B | 100 | 84.414 | Panthera_tigris_altaica |
| ENSG00000155846 | PPARGC1B | 50 | 49.550 | ENSPTIG00000007378 | PPARGC1A | 63 | 51.908 | Panthera_tigris_altaica |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSPTRG00000015954 | PPARGC1A | 60 | 51.908 | Pan_troglodytes |
| ENSG00000155846 | PPARGC1B | 100 | 99.316 | ENSPTRG00000017403 | PPARGC1B | 100 | 99.316 | Pan_troglodytes |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSPANG00000005420 | PPARGC1A | 57 | 51.908 | Papio_anubis |
| ENSG00000155846 | PPARGC1B | 100 | 96.285 | ENSPANG00000009512 | PPARGC1B | 100 | 96.285 | Papio_anubis |
| ENSG00000155846 | PPARGC1B | 99 | 51.622 | ENSPSIG00000008358 | PPARGC1B | 99 | 49.392 | Pelodiscus_sinensis |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSPEMG00000010892 | Ppargc1a | 55 | 51.145 | Peromyscus_maniculatus_bairdii |
| ENSG00000155846 | PPARGC1B | 100 | 78.850 | ENSPEMG00000018822 | Ppargc1b | 100 | 78.850 | Peromyscus_maniculatus_bairdii |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSPCIG00000015835 | PPARGC1A | 55 | 52.083 | Phascolarctos_cinereus |
| ENSG00000155846 | PPARGC1B | 90 | 59.823 | ENSPCIG00000020102 | PPARGC1B | 99 | 56.754 | Phascolarctos_cinereus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSPPYG00000014640 | PPARGC1A | 55 | 51.908 | Pongo_abelii |
| ENSG00000155846 | PPARGC1B | 100 | 98.729 | ENSPPYG00000015938 | PPARGC1B | 100 | 98.729 | Pongo_abelii |
| ENSG00000155846 | PPARGC1B | 72 | 82.171 | ENSPCAG00000012171 | PPARGC1B | 74 | 88.462 | Procavia_capensis |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSPCAG00000016308 | PPARGC1A | 55 | 51.908 | Procavia_capensis |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSPCOG00000022858 | PPARGC1A | 76 | 41.038 | Propithecus_coquereli |
| ENSG00000155846 | PPARGC1B | 100 | 87.402 | ENSPCOG00000024059 | PPARGC1B | 100 | 87.695 | Propithecus_coquereli |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSPVAG00000004965 | PPARGC1A | 55 | 51.145 | Pteropus_vampyrus |
| ENSG00000155846 | PPARGC1B | 100 | 83.934 | ENSPVAG00000001308 | PPARGC1B | 100 | 84.031 | Pteropus_vampyrus |
| ENSG00000155846 | PPARGC1B | 55 | 43.750 | ENSPNAG00000023871 | ppargc1b | 78 | 43.750 | Pygocentrus_nattereri |
| ENSG00000155846 | PPARGC1B | 51 | 54.198 | ENSPNAG00000010393 | ppargc1a | 65 | 54.198 | Pygocentrus_nattereri |
| ENSG00000155846 | PPARGC1B | 100 | 77.854 | ENSRNOG00000017503 | Ppargc1b | 100 | 77.854 | Rattus_norvegicus |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSRNOG00000004473 | Ppargc1a | 55 | 51.145 | Rattus_norvegicus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSRBIG00000032333 | PPARGC1A | 58 | 51.908 | Rhinopithecus_bieti |
| ENSG00000155846 | PPARGC1B | 100 | 95.431 | ENSRBIG00000036724 | PPARGC1B | 100 | 95.330 | Rhinopithecus_bieti |
| ENSG00000155846 | PPARGC1B | 99 | 95.178 | ENSRROG00000029594 | PPARGC1B | 99 | 93.606 | Rhinopithecus_roxellana |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSRROG00000010419 | PPARGC1A | 56 | 51.908 | Rhinopithecus_roxellana |
| ENSG00000155846 | PPARGC1B | 51 | 51.908 | ENSSBOG00000006423 | PPARGC1A | 56 | 51.908 | Saimiri_boliviensis_boliviensis |
| ENSG00000155846 | PPARGC1B | 100 | 92.773 | ENSSBOG00000028738 | PPARGC1B | 100 | 92.871 | Saimiri_boliviensis_boliviensis |
| ENSG00000155846 | PPARGC1B | 50 | 51.145 | ENSSHAG00000010033 | PPARGC1A | 53 | 52.083 | Sarcophilus_harrisii |
| ENSG00000155846 | PPARGC1B | 99 | 58.565 | ENSSHAG00000016400 | PPARGC1B | 99 | 58.565 | Sarcophilus_harrisii |
| ENSG00000155846 | PPARGC1B | 50 | 55.725 | ENSSMAG00000011571 | ppargc1a | 65 | 55.725 | Scophthalmus_maximus |
| ENSG00000155846 | PPARGC1B | 99 | 50.048 | ENSSPUG00000019271 | PPARGC1B | 99 | 49.285 | Sphenodon_punctatus |
| ENSG00000155846 | PPARGC1B | 53 | 53.435 | ENSSPUG00000013556 | PPARGC1A | 56 | 53.435 | Sphenodon_punctatus |
| ENSG00000155846 | PPARGC1B | 100 | 76.081 | ENSSSCG00000014437 | PPARGC1B | 100 | 76.465 | Sus_scrofa |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSSSCG00000029275 | PPARGC1A | 56 | 51.908 | Sus_scrofa |
| ENSG00000155846 | PPARGC1B | 99 | 48.204 | ENSTGUG00000000232 | PPARGC1B | 100 | 50.100 | Taeniopygia_guttata |
| ENSG00000155846 | PPARGC1B | 51 | 53.435 | ENSTGUG00000009487 | PPARGC1A | 56 | 53.435 | Taeniopygia_guttata |
| ENSG00000155846 | PPARGC1B | 51 | 48.739 | ENSTTRG00000005327 | PPARGC1A | 55 | 51.908 | Tursiops_truncatus |
| ENSG00000155846 | PPARGC1B | 99 | 83.778 | ENSTTRG00000008430 | PPARGC1B | 100 | 83.877 | Tursiops_truncatus |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSUMAG00000003557 | PPARGC1A | 56 | 51.908 | Ursus_maritimus |
| ENSG00000155846 | PPARGC1B | 99 | 83.299 | ENSUMAG00000005361 | PPARGC1B | 99 | 82.621 | Ursus_maritimus |
| ENSG00000155846 | PPARGC1B | 100 | 85.010 | ENSVVUG00000014358 | PPARGC1B | 100 | 84.816 | Vulpes_vulpes |
| ENSG00000155846 | PPARGC1B | 50 | 51.908 | ENSVVUG00000013365 | PPARGC1A | 55 | 51.908 | Vulpes_vulpes |
| ENSG00000155846 | PPARGC1B | 56 | 43.147 | ENSXETG00000020468 | ppargc1a | 53 | 51.908 | Xenopus_tropicalis |
| ENSG00000155846 | PPARGC1B | 90 | 45.217 | ENSXETG00000012711 | ppargc1b | 96 | 45.739 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001503 | ossification | - | IEA | Process |
| GO:0003712 | transcription coregulator activity | 21873635. | IBA | Function |
| GO:0003712 | transcription coregulator activity | 11854298. | IDA | Function |
| GO:0003723 | RNA binding | 11854298. | NAS | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 11854298. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0006355 | regulation of transcription, DNA-templated | 23836911. | IDA | Process |
| GO:0006390 | mitochondrial transcription | - | IEA | Process |
| GO:0007015 | actin filament organization | - | IEA | Process |
| GO:0008134 | transcription factor binding | 21873635. | IBA | Function |
| GO:0010694 | positive regulation of alkaline phosphatase activity | - | IEA | Process |
| GO:0016592 | mediator complex | 11854298. | IDA | Component |
| GO:0030331 | estrogen receptor binding | 11854298. | IDA | Function |
| GO:0030374 | nuclear receptor transcription coactivator activity | 21873635. | IBA | Function |
| GO:0030374 | nuclear receptor transcription coactivator activity | 11854298. | IDA | Function |
| GO:0030520 | intracellular estrogen receptor signaling pathway | 11854298. | IDA | Process |
| GO:0034614 | cellular response to reactive oxygen species | - | IEA | Process |
| GO:0042327 | positive regulation of phosphorylation | - | IEA | Process |
| GO:0045672 | positive regulation of osteoclast differentiation | - | IEA | Process |
| GO:0045780 | positive regulation of bone resorption | - | IEA | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | - | IEA | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 11854298. | IDA | Process |
| GO:0050682 | AF-2 domain binding | 11854298. | IPI | Function |
| GO:0051091 | positive regulation of DNA-binding transcription factor activity | 21873635. | IBA | Process |
| GO:0051384 | response to glucocorticoid | - | IEA | Process |
| GO:0051591 | response to cAMP | - | IEA | Process |
| GO:0060346 | bone trabecula formation | - | IEA | Process |
| GO:0120162 | positive regulation of cold-induced thermogenesis | 17090215. | ISS | Process |