
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| ESCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| ACC | ||
| HNSC | ||
| BLCA | ||
| LAML | ||
| UCEC | ||
| GBM | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000494253 | HK1-217 | 2848 | - | - | - (aa) | - | - |
| ENST00000483077 | HK1-214 | 750 | - | - | - (aa) | - | - |
| ENST00000464803 | HK1-208 | 567 | - | ENSP00000496531 | 59 (aa) | - | A0A2R8Y7T9 |
| ENST00000488644 | HK1-215 | 593 | - | - | - (aa) | - | - |
| ENST00000480047 | HK1-212 | 575 | - | - | - (aa) | - | - |
| ENST00000421088 | HK1-204 | 707 | - | ENSP00000398316 | 114 (aa) | - | B1AR62 |
| ENST00000360289 | HK1-203 | 3961 | XM_011539732 | ENSP00000353433 | 905 (aa) | XP_011538034 | P19367 |
| ENST00000643399 | HK1-218 | 3998 | - | ENSP00000494664 | 921 (aa) | - | P19367 |
| ENST00000436817 | HK1-205 | 3835 | - | ENSP00000415949 | 921 (aa) | - | P19367 |
| ENST00000448642 | HK1-206 | 3868 | XM_024447969 | ENSP00000402103 | 921 (aa) | XP_024303737 | P19367 |
| ENST00000359426 | HK1-202 | 3605 | XM_005269737 | ENSP00000352398 | 917 (aa) | XP_005269794 | P19367 |
| ENST00000479594 | HK1-211 | 680 | - | - | - (aa) | - | - |
| ENST00000450646 | HK1-207 | 449 | - | ENSP00000409761 | 30 (aa) | - | P78542 |
| ENST00000470050 | HK1-209 | 314 | - | - | - (aa) | - | - |
| ENST00000483054 | HK1-213 | 592 | - | - | - (aa) | - | - |
| ENST00000476368 | HK1-210 | 456 | - | ENSP00000495526 | 29 (aa) | - | A0A2R8YEE7 |
| ENST00000493591 | HK1-216 | 3745 | - | ENSP00000494917 | 92 (aa) | - | A0A2R8Y626 |
| ENST00000298649 | HK1-201 | 3229 | - | ENSP00000298649 | 916 (aa) | - | P19367 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00010 | Glycolysis / Gluconeogenesis | KEGG |
| hsa00051 | Fructose and mannose metabolism | KEGG |
| hsa00052 | Galactose metabolism | KEGG |
| hsa00500 | Starch and sucrose metabolism | KEGG |
| hsa00520 | Amino sugar and nucleotide sugar metabolism | KEGG |
| hsa00524 | Neomycin, kanamycin and gentamicin biosynthesis | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa01200 | Carbon metabolism | KEGG |
| hsa04066 | HIF-1 signaling pathway | KEGG |
| hsa04910 | Insulin signaling pathway | KEGG |
| hsa04930 | Type II diabetes mellitus | KEGG |
| hsa04973 | Carbohydrate digestion and absorption | KEGG |
| hsa05230 | Central carbon metabolism in cancer | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000156515 | rs2278745 | 10 | 69392335 | C | Iris color (b* coordinate) | 29109912 | [0.25-0.54] unit decrease | 0.394833 | EFO_0003949 |
| ENSG00000156515 | rs2278745 | 10 | 69392335 | C | Iris color (a* coordinate) | 29109912 | [0.25-0.54] unit decrease | 0.395 | EFO_0003949 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | High light scatter reticulocyte percentage of red cells | 27863252 | [0.077-0.099] unit increase | 0.08805049 | EFO_0007986 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Reticulocyte count | 27863252 | [0.11-0.13] unit increase | 0.1197591 | EFO_0007986 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Mean corpuscular hemoglobin | 27863252 | [0.068-0.09] unit increase | 0.07898236 | EFO_0004527 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Red blood cell count | 27863252 | [0.072-0.094] unit increase | 0.0833013 | EFO_0004305 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Mean corpuscular volume | 27863252 | [0.077-0.099] unit increase | 0.08811442 | EFO_0004526 |
| ENSG00000156515 | rs16926246 | 10 | 69333636 | ? | Hemoglobin levels | 28017375 | EFO_0004509 | ||
| ENSG00000156515 | rs10159477 | 10 | 69340132 | A | Red blood cell traits | 23222517 | [0.067-0.107] unit increase | 0.087 | EFO_0004509 |
| ENSG00000156515 | rs7072268 | 10 | 69340157 | A | Glycated hemoglobin levels | 19096518 | [NR] % increase | 0.05 | EFO_0004541 |
| ENSG00000156515 | rs16926246 | 10 | 69333636 | T | Hemoglobin | 19862010 | [0.08-0.14] g/dl increase | 0.11 | EFO_0004509 |
| ENSG00000156515 | rs16926246 | 10 | 69333636 | T | Hematocrit | 19862010 | [0.24-0.42] % increase | 0.33 | EFO_0004348 |
| ENSG00000156515 | rs4745982 | 10 | 69330087 | T | Glycated hemoglobin levels | 28898252 | [0.084-0.106] unit increase | 0.0954 | EFO_0004541 |
| ENSG00000156515 | rs10823343 | 10 | 69331257 | A | Glycated hemoglobin levels | 28898252 | [0.028-0.037] unit increase | 0.0325 | EFO_0004541 |
| ENSG00000156515 | rs10823343 | 10 | 69331257 | G | Hemoglobin A1c levels | 29844224 | [0.027-0.067] unit decrease | 0.047 | EFO_0007629 |
| ENSG00000156515 | rs11596587 | 10 | 69354232 | T | Ischemic stroke (cardioembolic) | 29531354 | [1.14-1.31] | 1.22 | EFO_1001976 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Hematocrit | 27863252 | [0.14-0.16] unit increase | 0.1517784 | EFO_0004348 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Hemoglobin concentration | 27863252 | [0.14-0.16] unit increase | 0.151392 | EFO_0004509 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Reticulocyte fraction of red cells | 27863252 | [0.092-0.115] unit increase | 0.1032995 | EFO_0007986 |
| ENSG00000156515 | rs72805692 | 10 | 69339353 | G | Immature fraction of reticulocytes | 27863252 | [0.027-0.048] unit increase | 0.03743759 | EFO_0007986 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | Red cell distribution width | 27863252 | [0.027-0.049] unit increase | 0.03797962 | EFO_0005192 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | C | High light scatter reticulocyte count | 27863252 | [0.09-0.112] unit increase | 0.1009026 | EFO_0007986 |
| ENSG00000156515 | rs16926246 | 10 | 69333636 | C | Glycated hemoglobin levels | 20858683 | [0.08-0.10] % increase | 0.09 | EFO_0004541 |
| ENSG00000156515 | rs17476364 | 10 | 69334748 | ? | Red cell distribution width | 30595370 | EFO_0005192 | ||
| ENSG00000156515 | rs17476364 | 10 | 69334748 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 | ||
| ENSG00000156515 | rs2305196 | 10 | 69384568 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 | ||
| ENSG00000156515 | rs17476364 | 10 | 69334748 | ? | Red blood cell count | 30595370 | EFO_0004305 |
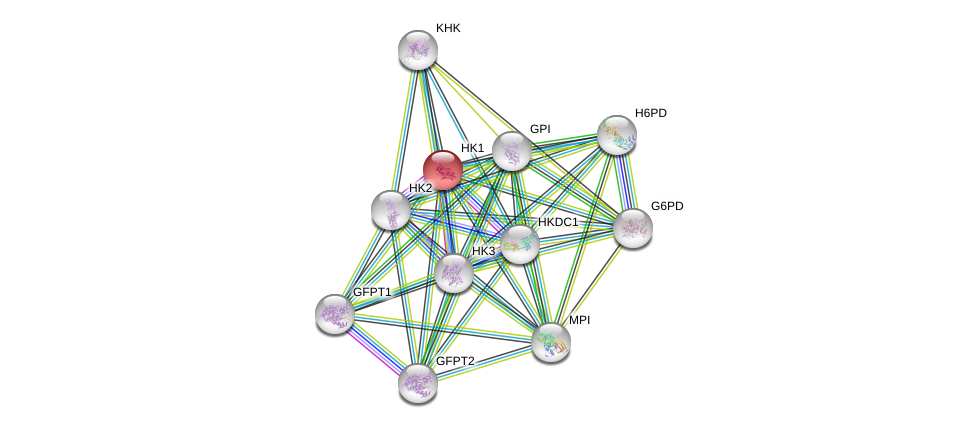
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000156515 | HK1 | 99 | 73.982 | ENSG00000159399 | HK2 | 99 | 73.982 | Homo_sapiens |
| ENSG00000156515 | HK1 | 99 | 73.982 | ENSMUSG00000000628 | Hk2 | 99 | 73.982 | Mus_musculus |
| ENSG00000156515 | HK1 | 94 | 35.857 | YGL253W | HXK2 | 98 | 35.438 | Saccharomyces_cerevisiae |
| ENSG00000156515 | HK1 | 99 | 34.447 | YFR053C | 97 | 34.447 | Saccharomyces_cerevisiae | |
| ENSG00000156515 | HK1 | 98 | 33.333 | YCL040W | GLK1 | 98 | 33.333 | Saccharomyces_cerevisiae |
| ENSG00000156515 | HK1 | 93 | 35.056 | YDR516C | 98 | 35.056 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001678 | cellular glucose homeostasis | 21873635. | IBA | Process |
| GO:0002741 | positive regulation of cytokine secretion involved in immune response | - | IEA | Process |
| GO:0004340 | glucokinase activity | 21873635. | IBA | Function |
| GO:0004396 | hexokinase activity | 27374331. | IMP | Function |
| GO:0005515 | protein binding | 28054552. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005536 | glucose binding | - | IEA | Function |
| GO:0005739 | mitochondrion | 20833797. | HDA | Component |
| GO:0005739 | mitochondrion | - | IDA | Component |
| GO:0005741 | mitochondrial outer membrane | - | IEA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006096 | glycolytic process | 21873635. | IBA | Process |
| GO:0008865 | fructokinase activity | 21873635. | IBA | Function |
| GO:0019158 | mannokinase activity | 21873635. | IBA | Function |
| GO:0042802 | identical protein binding | 28054552.29568061. | IPI | Function |
| GO:0042834 | peptidoglycan binding | 27374331. | IDA | Function |
| GO:0045121 | membrane raft | - | IEA | Component |
| GO:0046835 | carbohydrate phosphorylation | 27374331. | IMP | Process |
| GO:0050718 | positive regulation of interleukin-1 beta secretion | - | IEA | Process |
| GO:0051156 | glucose 6-phosphate metabolic process | - | IEA | Process |
| GO:0061621 | canonical glycolysis | - | TAS | Process |
| GO:0072655 | establishment of protein localization to mitochondrion | 23962723. | IMP | Process |
| GO:0072656 | maintenance of protein location in mitochondrion | 23962723. | IMP | Process |