
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LAML | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| KIRC | |||
| READ | |||
| TGCT | |||
| THCA | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| LUSC | ||
| BRCA | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| LGG |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 23229552 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000332070 | PHF6-201 | 4759 | - | ENSP00000329097 | 365 (aa) | - | Q8IWS0 |
| ENST00000370799 | PHF6-202 | 4186 | - | ENSP00000359835 | 324 (aa) | - | Q5JRC6 |
| ENST00000625464 | PHF6-205 | 4434 | - | ENSP00000487420 | 366 (aa) | - | A0A0D9SGE8 |
| ENST00000370803 | PHF6-204 | 4489 | - | ENSP00000359839 | 365 (aa) | - | Q8IWS0 |
| ENST00000370800 | PHF6-203 | 1150 | - | ENSP00000359836 | 312 (aa) | - | Q8IWS0 |
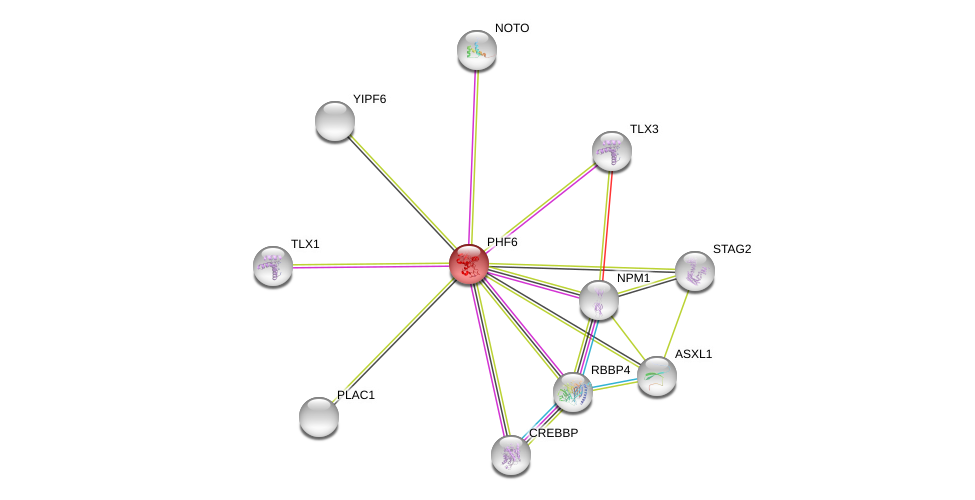
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 24554700. | IDA | Process |
| GO:0000777 | condensed chromosome kinetochore | - | IEA | Component |
| GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | 24554700. | IDA | Function |
| GO:0001835 | blastocyst hatching | - | IEA | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005515 | protein binding | 22720776. | IPI | Function |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005654 | nucleoplasm | 22720776. | IDA | Component |
| GO:0005730 | nucleolus | 22720776. | IDA | Component |
| GO:0015631 | tubulin binding | 22720776. | IPI | Function |
| GO:0019899 | enzyme binding | 22720776. | IPI | Function |
| GO:0042393 | histone binding | 22720776. | IPI | Function |
| GO:0042826 | histone deacetylase binding | 22720776. | IPI | Function |
| GO:0043021 | ribonucleoprotein complex binding | 22720776. | IPI | Function |
| GO:0043565 | sequence-specific DNA binding | 24554700. | IDA | Function |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0051219 | phosphoprotein binding | 22720776. | IPI | Function |
| GO:0097110 | scaffold protein binding | 22720776. | IPI | Function |