
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| SKCM | ||
| PRAD | ||
| BRCA | ||
| KIRP | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| DLBC | ||
| LUAD |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000486604 | CUL4B-206 | 1024 | - | - | - (aa) | - | - |
| ENST00000404115 | CUL4B-204 | 3264 | XM_005262481 | ENSP00000384109 | 913 (aa) | XP_005262538 | Q13620 |
| ENST00000336592 | CUL4B-201 | 3109 | XM_006724784 | ENSP00000338919 | 900 (aa) | XP_006724847 | K4DI93 |
| ENST00000371323 | CUL4B-203 | 846 | - | ENSP00000360374 | 234 (aa) | - | A6NE76 |
| ENST00000371322 | CUL4B-202 | 4902 | XM_011531399 | ENSP00000360373 | 895 (aa) | XP_011529701 | Q13620 |
| ENST00000467641 | CUL4B-205 | 836 | - | - | - (aa) | - | - |
| ENST00000497616 | CUL4B-207 | 1011 | - | - | - (aa) | - | - |
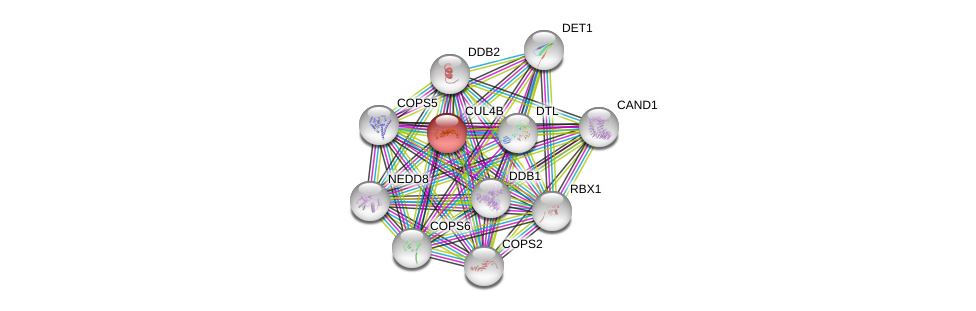
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000158290 | CUL4B | 100 | 97.442 | ENSMUSG00000031095 | Cul4b | 100 | 97.442 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000082 | G1/S transition of mitotic cell cycle | 8681378. | NAS | Process |
| GO:0000715 | nucleotide-excision repair, DNA damage recognition | - | TAS | Process |
| GO:0000717 | nucleotide-excision repair, DNA duplex unwinding | - | TAS | Process |
| GO:0003684 | damaged DNA binding | 22334663. | IDA | Function |
| GO:0005515 | protein binding | 12609982.16949367.16964240.17041588.18775313.19651607.21778237.22466964.22939624.23238014.25036637.25435324.25910212.26496610. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006283 | transcription-coupled nucleotide-excision repair | - | TAS | Process |
| GO:0006293 | nucleotide-excision repair, preincision complex stabilization | - | TAS | Process |
| GO:0006294 | nucleotide-excision repair, preincision complex assembly | - | TAS | Process |
| GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion | - | TAS | Process |
| GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion | - | TAS | Process |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 21873635. | IBA | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 21873635. | IBA | Process |
| GO:0010498 | proteasomal protein catabolic process | 25970626. | IDA | Process |
| GO:0010498 | proteasomal protein catabolic process | 25970626. | IMP | Process |
| GO:0016567 | protein ubiquitination | 28437394. | IMP | Process |
| GO:0019005 | SCF ubiquitin ligase complex | 21873635. | IBA | Component |
| GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 21873635. | IBA | Process |
| GO:0031175 | neuron projection development | - | IEA | Process |
| GO:0031461 | cullin-RING ubiquitin ligase complex | 21873635. | IBA | Component |
| GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | 21873635. | IBA | Component |
| GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | 18794347.21628527.22334663. | IDA | Component |
| GO:0031625 | ubiquitin protein ligase binding | 21873635. | IBA | Function |
| GO:0033683 | nucleotide-excision repair, DNA incision | - | TAS | Process |
| GO:0035518 | histone H2A monoubiquitination | 22334663. | IDA | Process |
| GO:0042769 | DNA damage response, detection of DNA damage | - | TAS | Process |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 21873635. | IBA | Process |
| GO:0043687 | post-translational protein modification | - | TAS | Process |
| GO:0045732 | positive regulation of protein catabolic process | - | IEA | Process |
| GO:0061630 | ubiquitin protein ligase activity | 21873635. | IBA | Function |
| GO:0070062 | extracellular exosome | 19056867. | HDA | Component |
| GO:0070911 | global genome nucleotide-excision repair | - | TAS | Process |
| GO:0070914 | UV-damage excision repair | 22334663. | IDA | Process |
| GO:0080008 | Cul4-RING E3 ubiquitin ligase complex | 21873635. | IBA | Component |
| GO:0080008 | Cul4-RING E3 ubiquitin ligase complex | 16949367. | IDA | Component |
| GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | - | IEA | Process |