
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| ESCA | |||
| KIRC | |||
| LGG | |||
| PAAD | |||
| READ | |||
| SKCM | |||
| TGCT | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| ACC | ||
| UCS | ||
| COAD | ||
| PAAD | ||
| BLCA | ||
| READ | ||
| LAML | ||
| GBM | ||
| LIHC | ||
| LUAD | ||
| UVM |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 31317183 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000508500 | LMNA-222 | 848 | - | ENSP00000424977 | 260 (aa) | - | A0A0C4DGC5 |
| ENST00000498722 | LMNA-217 | 1348 | - | - | - (aa) | - | - |
| ENST00000515824 | LMNA-225 | 393 | - | - | - (aa) | - | - |
| ENST00000504687 | LMNA-220 | 546 | - | ENSP00000426535 | 182 (aa) | - | H0YAB0 |
| ENST00000502751 | LMNA-219 | 570 | - | - | - (aa) | - | - |
| ENST00000469565 | LMNA-210 | 591 | - | - | - (aa) | - | - |
| ENST00000515711 | LMNA-224 | 569 | - | - | - (aa) | - | - |
| ENST00000470835 | LMNA-212 | 472 | - | - | - (aa) | - | - |
| ENST00000515459 | LMNA-223 | 757 | - | ENSP00000424518 | 38 (aa) | - | D6RB20 |
| ENST00000506981 | LMNA-221 | 576 | - | - | - (aa) | - | - |
| ENST00000473598 | LMNA-213 | 2015 | - | ENSP00000421821 | 565 (aa) | - | P02545 |
| ENST00000368301 | LMNA-207 | 2461 | - | ENSP00000357284 | 572 (aa) | - | P02545 |
| ENST00000368300 | LMNA-206 | 3190 | XM_011509534 | ENSP00000357283 | 664 (aa) | XP_011507836 | P02545 |
| ENST00000361308 | LMNA-202 | 1713 | - | ENSP00000355292 | 487 (aa) | - | Q3BDU5 |
| ENST00000478063 | LMNA-214 | 285 | - | - | - (aa) | - | - |
| ENST00000347559 | LMNA-201 | 3137 | - | ENSP00000292304 | 634 (aa) | - | P02545 |
| ENST00000448611 | LMNA-208 | 1943 | XM_011509533 | ENSP00000395597 | 574 (aa) | XP_011507835 | P02545 |
| ENST00000459904 | LMNA-209 | 916 | - | - | - (aa) | - | - |
| ENST00000496738 | LMNA-216 | 2405 | - | - | - (aa) | - | - |
| ENST00000470199 | LMNA-211 | 551 | - | - | - (aa) | - | - |
| ENST00000502357 | LMNA-218 | 537 | - | - | - (aa) | - | - |
| ENST00000495341 | LMNA-215 | 394 | - | - | - (aa) | - | - |
| ENST00000368297 | LMNA-203 | 1679 | - | ENSP00000357280 | 491 (aa) | - | Q5TCI8 |
| ENST00000368299 | LMNA-205 | 2253 | - | ENSP00000357282 | 614 (aa) | - | P02545 |
| ENST00000368298 | LMNA-204 | 1475 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000160789 | Platelet Function Tests | 1.3511000E-005 | - |
| ENSG00000160789 | Platelet Function Tests | 6.2900000E-006 | - |
| ENSG00000160789 | Platelet Function Tests | 8.6800000E-006 | - |
| ENSG00000160789 | Platelet Function Tests | 1.9100000E-006 | - |
| ENSG00000160789 | Platelet Function Tests | 9.7600000E-007 | - |
| ENSG00000160789 | Stroke | 3E-6 | 27997041 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000160789 | rs116480793 | 1 | 156108585 | T | Lung cancer in ever smokers | 28604730 | [1.153366335-1.439461007] | 1.2884976 | EFO_0006527|EFO_0001071 |
| ENSG00000160789 | rs58799304 | 1 | 156110653 | ? | Ischemic stroke | 27997041 | z score decrease | 4.644 | HP_0002140 |
| ENSG00000160789 | rs188625872 | 1 | 156114696 | C | Ovarian cancer | 30557369 | [2.17-6.49] | 3.7593985 | EFO_0001075 |
| ENSG00000160789 | rs112941217 | 1 | 156087626 | C | Ischemic heart disease in rheumatoid arthritis | 30251476 | [2.61-8.87] | 4.81 | EFO_0000685|EFO_0001425 |
| ENSG00000160789 | rs584025 | 1 | 156118735 | ? | White blood cell count | 30595370 | EFO_0004308 |
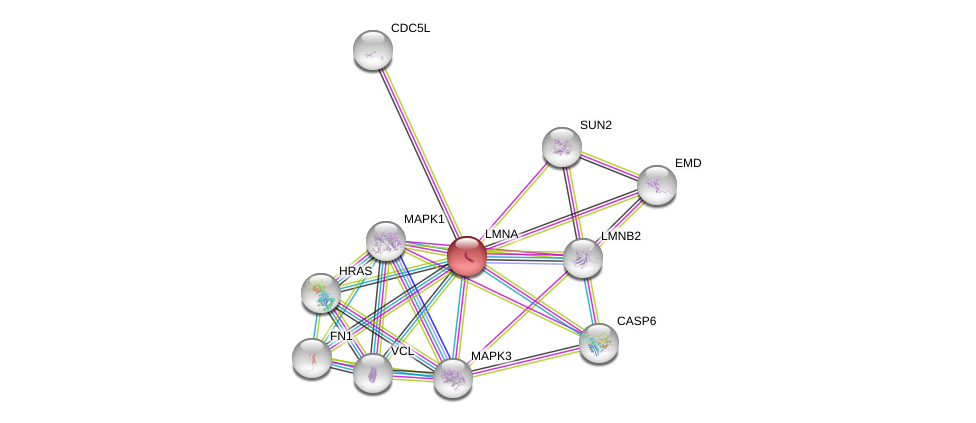
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000160789 | LMNA | 100 | 99.087 | ENSMUSG00000028063 | Lmna | 100 | 100.000 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005198 | structural molecule activity | - | IEA | Function |
| GO:0005515 | protein binding | 10514485.10727209.11801724.15140953.15161933.15671068.16247757.19323649.19933576.20000738.20580717.20618440.21418524.21949239.21988832.22399800.22555292.23658700.25416956.29568061.29997244.30021884. | IPI | Function |
| GO:0005634 | nucleus | 16791210. | HDA | Component |
| GO:0005634 | nucleus | 18809582.20810912. | IDA | Component |
| GO:0005635 | nuclear envelope | 18606848.27534416. | IDA | Component |
| GO:0005635 | nuclear envelope | 21610090. | IMP | Component |
| GO:0005635 | nuclear envelope | - | TAS | Component |
| GO:0005638 | lamin filament | - | IEA | Component |
| GO:0005654 | nucleoplasm | 27534416. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006606 | protein import into nucleus | - | IEA | Process |
| GO:0007084 | mitotic nuclear envelope reassembly | - | TAS | Process |
| GO:0008285 | negative regulation of cell proliferation | 27534416. | IMP | Process |
| GO:0010628 | positive regulation of gene expression | - | IEA | Process |
| GO:0016607 | nuclear speck | - | IEA | Component |
| GO:0030334 | regulation of cell migration | - | ISS | Process |
| GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | - | ISS | Process |
| GO:0031647 | regulation of protein stability | - | IEA | Process |
| GO:0031965 | nuclear membrane | 16791210. | HDA | Component |
| GO:0034504 | protein localization to nucleus | - | ISS | Process |
| GO:0036498 | IRE1-mediated unfolded protein response | - | TAS | Process |
| GO:0055015 | ventricular cardiac muscle cell development | - | IEA | Process |
| GO:0071456 | cellular response to hypoxia | 20810912. | IEP | Process |
| GO:0072201 | negative regulation of mesenchymal cell proliferation | - | IEA | Process |
| GO:0090201 | negative regulation of release of cytochrome c from mitochondria | - | IEA | Process |
| GO:0090343 | positive regulation of cell aging | 20458013. | IDA | Process |
| GO:1900114 | positive regulation of histone H3-K9 trimethylation | - | IEA | Process |
| GO:1900180 | regulation of protein localization to nucleus | - | IEA | Process |
| GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | - | ISS | Process |
| GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | - | IEA | Process |