
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| READ | |||
| STAD |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| SARC | ||
| SKCM | ||
| BRCA | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| READ | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000551876 | RACGAP1-224 | 539 | - | ENSP00000449186 | 130 (aa) | - | F8VWX0 |
| ENST00000548598 | RACGAP1-213 | 582 | - | ENSP00000449574 | 49 (aa) | - | F8VVY0 |
| ENST00000552004 | RACGAP1-225 | 530 | - | ENSP00000448136 | 107 (aa) | - | F8VUW9 |
| ENST00000546595 | RACGAP1-204 | 634 | - | ENSP00000449963 | 152 (aa) | - | F8VV37 |
| ENST00000427314 | RACGAP1-202 | 3229 | - | ENSP00000404190 | 632 (aa) | - | Q9H0H5 |
| ENST00000551260 | RACGAP1-223 | 555 | - | ENSP00000448860 | 56 (aa) | - | F8VXH1 |
| ENST00000552921 | RACGAP1-228 | 563 | - | ENSP00000447393 | 150 (aa) | - | F8VZ66 |
| ENST00000548824 | RACGAP1-215 | 586 | - | ENSP00000449170 | 174 (aa) | - | F8VWY4 |
| ENST00000551016 | RACGAP1-221 | 2193 | XM_011538238 | ENSP00000449374 | 632 (aa) | XP_011536540 | Q9H0H5 |
| ENST00000548961 | RACGAP1-216 | 387 | - | ENSP00000446889 | 117 (aa) | - | F8VZ41 |
| ENST00000548644 | RACGAP1-214 | 624 | - | ENSP00000449620 | 177 (aa) | - | F8VVE5 |
| ENST00000552310 | RACGAP1-227 | 932 | - | ENSP00000448697 | 293 (aa) | - | F8VRD2 |
| ENST00000552157 | RACGAP1-226 | 386 | - | ENSP00000448968 | 83 (aa) | - | F8VYH6 |
| ENST00000550149 | RACGAP1-219 | 570 | - | ENSP00000446642 | 155 (aa) | - | F8W1E5 |
| ENST00000550651 | RACGAP1-220 | 551 | - | ENSP00000449959 | 111 (aa) | - | F8VV39 |
| ENST00000548158 | RACGAP1-210 | 596 | - | - | - (aa) | - | - |
| ENST00000548320 | RACGAP1-212 | 554 | - | ENSP00000448507 | 34 (aa) | - | F8VRL2 |
| ENST00000548247 | RACGAP1-211 | 564 | - | ENSP00000450074 | 67 (aa) | - | F8VQF5 |
| ENST00000549777 | RACGAP1-218 | 570 | - | ENSP00000448707 | 129 (aa) | - | F8VS54 |
| ENST00000547905 | RACGAP1-209 | 2190 | - | ENSP00000449370 | 632 (aa) | - | Q9H0H5 |
| ENST00000546723 | RACGAP1-205 | 574 | - | ENSP00000449669 | 155 (aa) | - | F8VV47 |
| ENST00000454520 | RACGAP1-203 | 3080 | XM_017019221 | ENSP00000404808 | 632 (aa) | XP_016874710 | Q9H0H5 |
| ENST00000546764 | RACGAP1-206 | 581 | - | ENSP00000447177 | 139 (aa) | - | F8W0L1 |
| ENST00000551145 | RACGAP1-222 | 584 | - | ENSP00000450064 | 124 (aa) | - | F8VQZ5 |
| ENST00000549342 | RACGAP1-217 | 841 | - | ENSP00000449565 | 280 (aa) | - | H0YIK5 |
| ENST00000546786 | RACGAP1-207 | 568 | - | ENSP00000447429 | 136 (aa) | - | F8W1T4 |
| ENST00000547061 | RACGAP1-208 | 581 | - | - | - (aa) | - | - |
| ENST00000312377 | RACGAP1-201 | 3073 | XM_006719359 | ENSP00000309871 | 632 (aa) | XP_006719422 | Q9H0H5 |
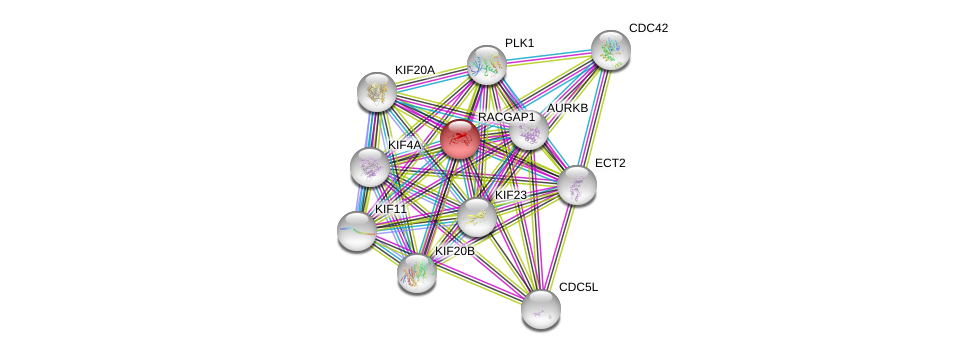
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000281 | mitotic cytokinesis | 11085985.19468302. | IDA | Process |
| GO:0000281 | mitotic cytokinesis | 16236794.23235882. | IMP | Process |
| GO:0000915 | actomyosin contractile ring assembly | 16129829. | IMP | Process |
| GO:0001669 | acrosomal vesicle | - | IEA | Component |
| GO:0005096 | GTPase activator activity | 10979956. | IDA | Function |
| GO:0005096 | GTPase activator activity | 15642749. | IMP | Function |
| GO:0005515 | protein binding | 11278976.11782313.12590651.12689593.14744859.16103226.16129829.16236794.18201571.18511905.19468300.19468302.22580824.22750944.25068414.25074804.25416956.26496610. | IPI | Function |
| GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding | 23235882. | IDA | Function |
| GO:0005634 | nucleus | 11085985.11782313.16103226. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005874 | microtubule | - | IEA | Component |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | - | TAS | Process |
| GO:0007018 | microtubule-based movement | - | TAS | Process |
| GO:0007283 | spermatogenesis | 12590651. | IEP | Process |
| GO:0007405 | neuroblast proliferation | - | ISS | Process |
| GO:0008017 | microtubule binding | 11782313. | IDA | Function |
| GO:0008272 | sulfate transport | 11278976. | IDA | Process |
| GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | - | TAS | Process |
| GO:0019901 | protein kinase binding | 19468302. | IPI | Function |
| GO:0030496 | midbody | 11782313.16103226.18511905. | IDA | Component |
| GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | 23235882. | IDA | Component |
| GO:0032154 | cleavage furrow | 16103226. | IDA | Component |
| GO:0032467 | positive regulation of cytokinesis | 16103226. | IDA | Process |
| GO:0032467 | positive regulation of cytokinesis | 11782313.15642749. | IMP | Process |
| GO:0035556 | intracellular signal transduction | - | IEA | Process |
| GO:0043014 | alpha-tubulin binding | 11085985. | IDA | Function |
| GO:0043015 | gamma-tubulin binding | 11085985. | IDA | Function |
| GO:0043547 | positive regulation of GTPase activity | - | IEA | Process |
| GO:0045995 | regulation of embryonic development | - | ISS | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0048487 | beta-tubulin binding | 11085985. | IDA | Function |
| GO:0051056 | regulation of small GTPase mediated signal transduction | - | TAS | Process |
| GO:0051233 | spindle midzone | 19468302. | IDA | Component |
| GO:0051256 | mitotic spindle midzone assembly | 16103226. | IDA | Process |
| GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 15642749. | IMP | Process |
| GO:0070062 | extracellular exosome | 19199708. | HDA | Component |
| GO:0072686 | mitotic spindle | 16103226. | IDA | Component |
| GO:0072686 | mitotic spindle | 15642749. | IMP | Component |
| GO:0090543 | Flemming body | - | IEA | Component |
| GO:0097149 | centralspindlin complex | 11782313.16236794. | IDA | Component |