
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| LIHC | |||
| LUAD | |||
| MESO |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| SARC | ||
| SKCM | ||
| KIRP | ||
| PAAD | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LGG | ||
| LUAD |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 27289039 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000511767 | PDLIM5-216 | 463 | - | - | - (aa) | - | - |
| ENST00000359265 | PDLIM5-203 | 1189 | - | ENSP00000352210 | 136 (aa) | - | Q96HC4 |
| ENST00000380180 | PDLIM5-205 | 2020 | - | ENSP00000369527 | 234 (aa) | - | Q96HC4 |
| ENST00000508216 | PDLIM5-210 | 1794 | XM_024453877 | ENSP00000426804 | 234 (aa) | XP_024309645 | Q96HC4 |
| ENST00000318007 | PDLIM5-202 | 1960 | - | ENSP00000322021 | 214 (aa) | - | Q96HC4 |
| ENST00000542407 | PDLIM5-221 | 2979 | XM_005262698 | ENSP00000442187 | 487 (aa) | XP_005262755 | Q96HC4 |
| ENST00000627587 | PDLIM5-223 | 1853 | - | ENSP00000486938 | 39 (aa) | - | A0A0D9SFW3 |
| ENST00000504489 | PDLIM5-208 | 538 | - | ENSP00000423009 | 38 (aa) | - | D6RAA1 |
| ENST00000380176 | PDLIM5-204 | 3098 | - | - | - (aa) | - | - |
| ENST00000509357 | PDLIM5-213 | 554 | - | ENSP00000422833 | 85 (aa) | - | H0Y929 |
| ENST00000509333 | PDLIM5-212 | 743 | - | - | - (aa) | - | - |
| ENST00000514743 | PDLIM5-219 | 3356 | - | ENSP00000424360 | 625 (aa) | - | Q96HC4 |
| ENST00000508531 | PDLIM5-211 | 480 | - | - | - (aa) | - | - |
| ENST00000506632 | PDLIM5-209 | 907 | - | ENSP00000422528 | 231 (aa) | - | H0Y8Y3 |
| ENST00000512274 | PDLIM5-217 | 372 | - | ENSP00000426379 | 47 (aa) | - | D6RGG6 |
| ENST00000437932 | PDLIM5-206 | 5713 | - | ENSP00000398469 | 271 (aa) | - | A0A0A0MSP3 |
| ENST00000514830 | PDLIM5-220 | 802 | - | - | - (aa) | - | - |
| ENST00000510099 | PDLIM5-214 | 554 | - | ENSP00000486834 | 176 (aa) | - | A0A0D9SFR4 |
| ENST00000615540 | PDLIM5-222 | 6219 | XM_005262693 | ENSP00000480359 | 625 (aa) | XP_005262750 | Q96HC4 |
| ENST00000317968 | PDLIM5-201 | 6083 | XM_006714069 | ENSP00000321746 | 596 (aa) | XP_006714132 | Q96HC4 |
| ENST00000511586 | PDLIM5-215 | 547 | - | - | - (aa) | - | - |
| ENST00000503974 | PDLIM5-207 | 2081 | - | ENSP00000424297 | 483 (aa) | - | Q96HC4 |
| ENST00000513341 | PDLIM5-218 | 794 | - | ENSP00000429577 | 196 (aa) | - | H0YBI4 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000163110 | Cholesterol | 1.71584995678021E-6 | 17903299 |
| ENSG00000163110 | Cholesterol | 2.03694257139506E-6 | 17903299 |
| ENSG00000163110 | Cholesterol, LDL | 3.63075972197746E-7 | 17903299 |
| ENSG00000163110 | Cholesterol, LDL | 5.44259226958239E-9 | 17903299 |
| ENSG00000163110 | Parietal Lobe | 6.20174811485352E-5 | 17903297 |
| ENSG00000163110 | Parietal Lobe | 3.09990700411156E-5 | 17903297 |
| ENSG00000163110 | Parietal Lobe | 8.52998395484139E-5 | 17903297 |
| ENSG00000163110 | Citalopram | 5.0800000E-005 | - |
| ENSG00000163110 | Citalopram | 9.2800000E-005 | - |
| ENSG00000163110 | Myocardial Infarction | 2.1840000E-005 | - |
| ENSG00000163110 | Myocardial Infarction | 1.8900000E-005 | - |
| ENSG00000163110 | Myocardial Infarction | 2.1090000E-005 | - |
| ENSG00000163110 | Myocardial Infarction | 1.8790000E-005 | - |
| ENSG00000163110 | Myocardial Infarction | 1.7660000E-005 | - |
| ENSG00000163110 | Myocardial Infarction | 1.2750000E-005 | - |
| ENSG00000163110 | Arterial Pressure | 6.5936251E-005 | - |
| ENSG00000163110 | Prostatic Neoplasms | 5E-6 | 22219177 |
| ENSG00000163110 | Amyotrophic lateral sclerosis 1 | 6E-6 | 24529757 |
| ENSG00000163110 | PCSK9 protein, human | 5E-6 | 25089948 |
| ENSG00000163110 | Simvastatin | 5E-6 | 25089948 |
| ENSG00000163110 | Pancreatitis | 3E-7 | 27114598 |
| ENSG00000163110 | Precursor Cell Lymphoblastic Leukemia-Lymphoma | 3E-7 | 27114598 |
| ENSG00000163110 | Electrocardiography | 6E-6 | 23166209 |
| ENSG00000163110 | Adipose Tissue | 9E-6 | 22589738 |
| ENSG00000163110 | Estradiol | 5E-7 | 23518928 |
| ENSG00000163110 | Lewy Body Disease | 8E-6 | 25188341 |
| ENSG00000163110 | Prostatic Neoplasms | 4E-15 | 19767753 |
| ENSG00000163110 | Prostatic Neoplasms | 1E-11 | 19767753 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000163110 | rs11731606 | 4 | 94534108 | T | Mean platelet volume | 27863252 | [0.017-0.035] unit decrease | 0.0259829 | EFO_0004584 |
| ENSG00000163110 | rs200495769 | 4 | 94573377 | T | Asparaginase-induced acute pancreatitis in acute lymphoblastic leukemia (onset time) | 27114598 | 5.7 | EFO_1001507|EFO_0000220 | |
| ENSG00000163110 | rs17021918 | 4 | 94641726 | ? | Prostate cancer | 19767753 | [1.08-1.15] | 1.11 | EFO_0001663 |
| ENSG00000163110 | rs12500426 | 4 | 94593458 | ? | Prostate cancer | 19767753 | [1.05-1.12] | 1.08 | EFO_0001663 |
| ENSG00000163110 | rs2452600 | 4 | 94575731 | T | Coronary artery disease | 29212778 | [0.023-0.05] unit decrease | 0.0365 | EFO_0000378 |
| ENSG00000163110 | rs17336353 | 4 | 94595365 | G | Response to simvastatin treatment (PCSK9 protein level change) | 25089948 | [NR] unit increase | 0.151 | GO_1903491|EFO_0006899 |
| ENSG00000163110 | rs2452591 | 4 | 94555568 | A | Lewy body disease | 25188341 | [0.15-0.39] unit decrease | 0.2693 | EFO_0006792|EFO_0006799 |
| ENSG00000163110 | rs71581597 | 4 | 94636417 | ? | Heel bone mineral density | 30048462 | [0.014-0.024] unit increase | 0.0188892 | EFO_0009270 |
| ENSG00000163110 | rs12511169 | 4 | 94652847 | T | Pulse pressure | 30578418 | [0.096-0.174] mmHg increase | 0.1351 | EFO_0005763 |
| ENSG00000163110 | rs17021918 | 4 | 94641726 | C | Prostate cancer | 29892016 | [1.07-1.11] | 1.09 | EFO_0001663 |
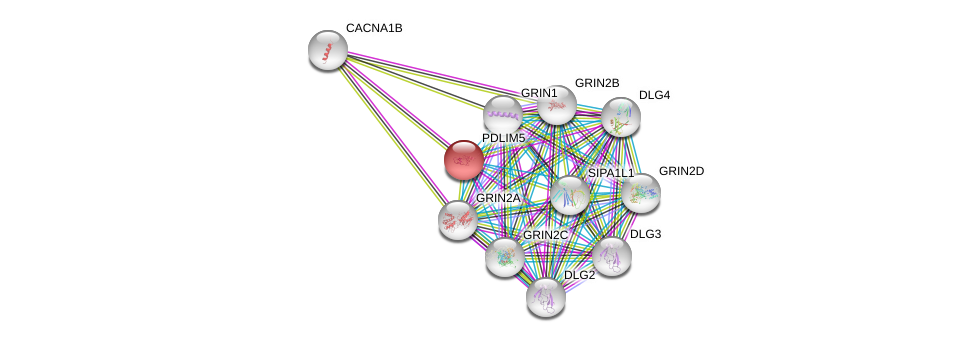
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000163110 | PDLIM5 | 99 | 95.833 | ENSMUSG00000028273 | Pdlim5 | 100 | 94.393 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003779 | actin binding | - | ISS | Function |
| GO:0005080 | protein kinase C binding | - | ISS | Function |
| GO:0005515 | protein binding | 16549780.25416956. | IPI | Function |
| GO:0005829 | cytosol | - | ISS | Component |
| GO:0005913 | cell-cell adherens junction | 25468996. | HDA | Component |
| GO:0014069 | postsynaptic density | - | ISS | Component |
| GO:0015629 | actin cytoskeleton | - | IDA | Component |
| GO:0016020 | membrane | - | ISS | Component |
| GO:0042805 | actinin binding | - | ISS | Function |
| GO:0043005 | neuron projection | - | IEA | Component |
| GO:0045211 | postsynaptic membrane | - | IEA | Component |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0047485 | protein N-terminus binding | - | IEA | Function |
| GO:0051963 | regulation of synapse assembly | - | ISS | Process |
| GO:0061001 | regulation of dendritic spine morphogenesis | - | ISS | Process |
| GO:0061049 | cell growth involved in cardiac muscle cell development | - | ISS | Process |
| GO:0098609 | cell-cell adhesion | - | IEA | Process |
| GO:0098641 | cadherin binding involved in cell-cell adhesion | 25468996. | HDA | Function |