
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| LUAD | |||
| MESO | |||
| PRAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| MESO | ||
| HNSC | ||
| LUSC | ||
| ESCA | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| THCA | ||
| LUAD |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000511552 | ANXA5-206 | 1036 | - | - | - (aa) | - | - |
| ENST00000501272 | ANXA5-202 | 1363 | - | ENSP00000424106 | 260 (aa) | - | D6RBL5 |
| ENST00000513523 | ANXA5-208 | 582 | - | - | - (aa) | - | - |
| ENST00000506395 | ANXA5-203 | 1606 | - | ENSP00000421421 | 163 (aa) | - | E9PHT9 |
| ENST00000509648 | ANXA5-205 | 1083 | - | - | - (aa) | - | - |
| ENST00000515017 | ANXA5-210 | 1190 | - | ENSP00000424199 | 220 (aa) | - | D6RBE9 |
| ENST00000513728 | ANXA5-209 | 447 | - | ENSP00000427135 | 35 (aa) | - | D6RCN3 |
| ENST00000515717 | ANXA5-211 | 594 | - | - | - (aa) | - | - |
| ENST00000509016 | ANXA5-204 | 556 | - | - | - (aa) | - | - |
| ENST00000296511 | ANXA5-201 | 1762 | - | ENSP00000296511 | 320 (aa) | - | P08758 |
| ENST00000513428 | ANXA5-207 | 868 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000164111 | rs55645543 | 4 | 121686023 | C | Age at smoking initiation in chronic obstructive pulmonary disease | 21685187 | unit increase | 0.228 | EFO_0000341|EFO_0005670 |
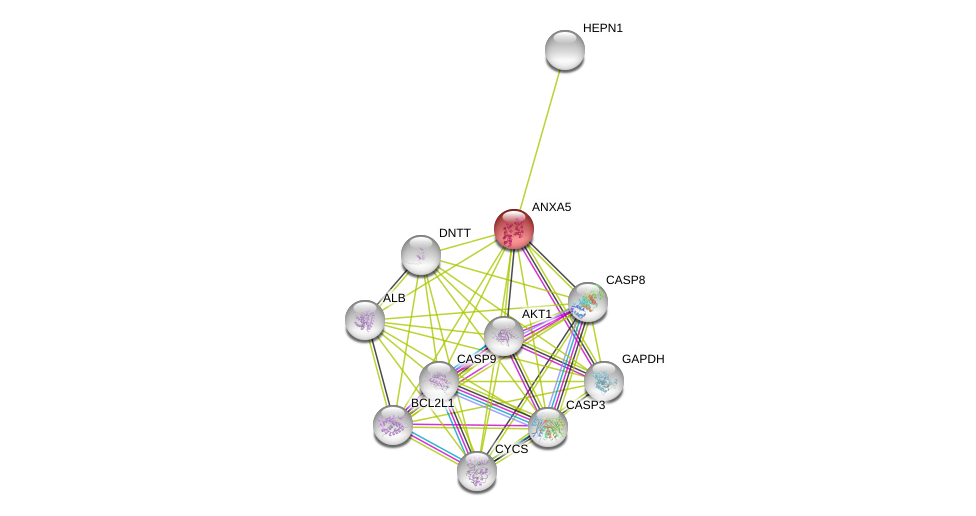
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002576 | platelet degranulation | - | TAS | Process |
| GO:0004859 | phospholipase inhibitor activity | 2967495. | TAS | Function |
| GO:0005388 | calcium-transporting ATPase activity | 21873635. | IBA | Function |
| GO:0005509 | calcium ion binding | - | IEA | Function |
| GO:0005515 | protein binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 21044950. | IPI | Function |
| GO:0005543 | phospholipid binding | 2967291. | TAS | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 21873635. | IBA | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 2138016. | IDA | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005737 | cytoplasm | 16130169. | TAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0007165 | signal transduction | 16130169. | TAS | Process |
| GO:0007596 | blood coagulation | - | IEA | Process |
| GO:0008201 | heparin binding | 21873635. | IBA | Function |
| GO:0009897 | external side of plasma membrane | - | IEA | Component |
| GO:0010033 | response to organic substance | 21873635. | IBA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016020 | membrane | 21873635. | IBA | Component |
| GO:0017046 | peptide hormone binding | 21873635. | IBA | Function |
| GO:0030195 | negative regulation of blood coagulation | 21873635. | IBA | Process |
| GO:0030971 | receptor tyrosine kinase binding | 21873635. | IBA | Function |
| GO:0043065 | positive regulation of apoptotic process | 21873635. | IBA | Process |
| GO:0043066 | negative regulation of apoptotic process | 16130169. | TAS | Process |
| GO:0043086 | negative regulation of catalytic activity | - | IEA | Process |
| GO:0051260 | protein homooligomerization | 21873635. | IBA | Process |
| GO:0051283 | negative regulation of sequestering of calcium ion | 21873635. | IBA | Process |
| GO:0051592 | response to calcium ion | 21873635. | IBA | Process |
| GO:0060090 | molecular adaptor activity | 21873635. | IBA | Function |
| GO:0062023 | collagen-containing extracellular matrix | 28675934. | HDA | Component |
| GO:0062023 | collagen-containing extracellular matrix | 28327460. | HDA | Component |
| GO:0070062 | extracellular exosome | 19056867.20458337.21362503.23533145. | HDA | Component |
| GO:0070588 | calcium ion transmembrane transport | - | IEA | Process |
| GO:0071284 | cellular response to lead ion | 21873635. | IBA | Process |
| GO:0072563 | endothelial microparticle | - | IEA | Component |
| GO:0097066 | response to thyroid hormone | 21873635. | IBA | Process |
| GO:0097211 | cellular response to gonadotropin-releasing hormone | 21873635. | IBA | Process |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | - | IEA | Process |
| GO:1901317 | regulation of flagellated sperm motility | 21873635. | IBA | Process |
| GO:1902721 | negative regulation of prolactin secretion | 21873635. | IBA | Process |