
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| CHOL | |||
| ESCA | |||
| LUAD | |||
| MESO | |||
| PAAD | |||
| SARC | |||
| SKCM | |||
| STAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| MESO | ||
| SKCM | ||
| COAD | ||
| PAAD | ||
| LAML | ||
| GBM | ||
| LIHC | ||
| LUAD | ||
| UVM |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 25752600 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000370318 | CGAS-202 | 1917 | - | ENSP00000359342 | 447 (aa) | - | Q8N884 |
| ENST00000459924 | CGAS-203 | 861 | - | - | - (aa) | - | - |
| ENST00000370315 | CGAS-201 | 3182 | - | ENSP00000359339 | 522 (aa) | - | Q8N884 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000164430 | Blood Pressure | 7.8560000E-005 | - |
| ENSG00000164430 | Creatinine | 7.2950000E-006 | - |
| ENSG00000164430 | Creatinine | 5.0790000E-006 | - |
| ENSG00000164430 | 3-hydroxy-1-methylpropylmercapturic acid | 4E-7 | 26053186 |
| ENSG00000164430 | Smoking | 4E-7 | 26053186 |
| ENSG00000164430 | S-(3-hydroxypropyl)cysteine N-acetate | 5E-6 | 26053186 |
| ENSG00000164430 | Smoking | 5E-6 | 26053186 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000164430 | rs311671 | 6 | 73429655 | ? | 3-hydroxypropylmercapturic acid levels in smokers | 26053186 | [NR] unit decrease | 0.493 | EFO_0004318|EFO_0007014 |
| ENSG00000164430 | rs311671 | 6 | 73429655 | ? | 3-hydroxy-1-methylpropylmercapturic acid levels in smokers | 26053186 | [NR] unit decrease | 0.4928 | EFO_0004318|EFO_0007015 |
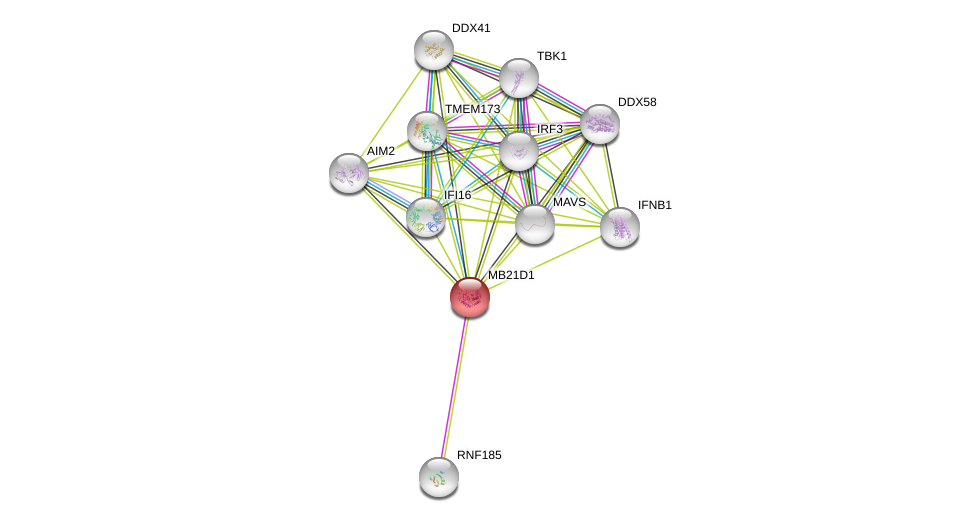
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002218 | activation of innate immune response | 23929945.24077100.26046437.28363908.30007416. | IDA | Process |
| GO:0002218 | activation of innate immune response | 23258413. | IMP | Process |
| GO:0002230 | positive regulation of defense response to virus by host | 23929945.24077100.26046437. | IDA | Process |
| GO:0002230 | positive regulation of defense response to virus by host | 23258413. | IMP | Process |
| GO:0002637 | regulation of immunoglobulin production | - | IEA | Process |
| GO:0003677 | DNA binding | 23258413. | IDA | Function |
| GO:0003682 | chromatin binding | 28738408. | IDA | Function |
| GO:0003690 | double-stranded DNA binding | 24077100.28363908.29937271.30007416. | IDA | Function |
| GO:0005515 | protein binding | 26046437.30135424. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005634 | nucleus | 29937271.30356214. | IDA | Component |
| GO:0005829 | cytosol | 23258413.29937271.30356214. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006281 | DNA repair | - | IEA | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 28738408. | IDA | Process |
| GO:0008340 | determination of adult lifespan | - | IEA | Process |
| GO:0010753 | positive regulation of cGMP-mediated signaling | 23258413.24077100.28738408. | IDA | Process |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0032481 | positive regulation of type I interferon production | 24077100.26046437.28363908.28738408. | IDA | Process |
| GO:0032481 | positive regulation of type I interferon production | - | TAS | Process |
| GO:0035861 | site of double-strand break | 30356214. | IDA | Component |
| GO:0038001 | paracrine signaling | 24077100. | IDA | Process |
| GO:0043950 | positive regulation of cAMP-mediated signaling | 23258413.24077100.28738408.30007416. | IDA | Process |
| GO:0045087 | innate immune response | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0050863 | regulation of T cell activation | - | IEA | Process |
| GO:0051607 | defense response to virus | 21478870.23929945.24077100.26046437. | IDA | Process |
| GO:0061501 | cyclic-GMP-AMP synthase activity | 23258412. | EXP | Function |
| GO:0061501 | cyclic-GMP-AMP synthase activity | 23258413.24077100.26300263.28214358.28363908.28738408.28934246.29937271.30007416. | IDA | Function |
| GO:0071360 | cellular response to exogenous dsRNA | 23258413.24077100.28214358.28363908. | IDA | Process |
| GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 30356214. | IDA | Process |
| GO:2000774 | positive regulation of cellular senescence | - | ISS | Process |