
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| DLBC | |||
| ESCA | |||
| PAAD | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| MESO | ||
| SKCM | ||
| KIRP | ||
| COAD | ||
| READ | ||
| LAML | ||
| KICH | ||
| LIHC | ||
| LGG | ||
| THCA |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000298139 | HRDC | PF00570.23 | 1.5e-11 | 1 | 1 |
| ENSP00000298139 | DEAD | PF00270.29 | 3.5e-15 | 1 | 1 |
| ENSP00000298139 | DNA_pol_A_exo1 | PF01612.20 | 1.2e-35 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000298139 | WRN-201 | 5215 | XM_011544639 | ENSP00000298139 | 1432 (aa) | XP_011542941 | Q14191 |
| ENST00000520169 | WRN-202 | 629 | - | - | - (aa) | - | - |
| ENST00000521620 | WRN-203 | 3593 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000165392 | Exercise Test | 6.31966375083062E-6 | 17903301 |
| ENSG00000165392 | Iron | 2.8850000E-004 | 19880490 |
| ENSG00000165392 | Iron | 3.1610000E-004 | 19880490 |
| ENSG00000165392 | Iron | 9.2310000E-004 | 19880490 |
| ENSG00000165392 | Iron | 8.2240000E-004 | 19880490 |
| ENSG00000165392 | Iron | 6.7920000E-004 | 19880490 |
| ENSG00000165392 | Waist-Hip Ratio | 7.8790000E-005 | - |
| ENSG00000165392 | Waist-Hip Ratio | 9.2880000E-005 | - |
| ENSG00000165392 | Angiotensin-Converting Enzyme Inhibitors | 2E-6 | 28084903 |
| ENSG00000165392 | Cough | 2E-6 | 28084903 |
| ENSG00000165392 | Immunoglobulins | 3E-6 | 23382691 |
| ENSG00000165392 | Longevity | 6E-8 | 25918517 |
| ENSG00000165392 | Immunoglobulins | 1E-6 | 23382691 |
| ENSG00000165392 | Exercise Test | 6E-6 | 17903301 |
| ENSG00000165392 | Immunoglobulins | 1E-6 | 23382691 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000165392 | rs10954779 | 8 | 31162081 | T | Intelligence (MTAG) | 29326435 | [0.011-0.023] unit decrease | 0.017427584 | EFO_0004337 |
| ENSG00000165392 | rs2737339 | 8 | 31155954 | A | Cognitive performance | 30038396 | [0.013-0.024] unit decrease | 0.0183 | EFO_0008354 |
| ENSG00000165392 | rs62506104 | 8 | 31162492 | A | Educational attainment (MTAG) | 30038396 | [0.006-0.0126] unit decrease | 0.0093 | EFO_0004784 |
| ENSG00000165392 | rs2553268 | 8 | 31078840 | ? | Exercise treadmill test traits | 17903301 | EFO_0004328 | ||
| ENSG00000165392 | rs11574358 | 8 | 31147066 | ? | Lifespan | 25918517 | [NR] unit increase | 5.584 | EFO_0004300 |
| ENSG00000165392 | rs117173564 | 8 | 31153953 | ? | Cough in response to angiotensin-converting enzyme inhibitor drugs | 28084903 | [2.88-12.82] | 6.07 | EFO_0005325|HP_0012735 |
| ENSG00000165392 | rs2737339 | 8 | 31155954 | A | Cognitive performance (MTAG) | 30038396 | [0.013-0.022] unit decrease | 0.0177 | EFO_0008354 |
| ENSG00000165392 | rs16877794 | 8 | 31158766 | ? | General cognitive ability | 29844566 | z-score increase | 5.212 | EFO_0004337 |
| ENSG00000165392 | rs10954779 | 8 | 31162081 | T | Intelligence | 29942086 | z-score decrease | 5.93 | EFO_0004337 |
| ENSG00000165392 | rs41520844 | 8 | 31135670 | ? | Adolescent idiopathic scoliosis | 30019117 | EFO_0005423 | ||
| ENSG00000165392 | rs62506074 | 8 | 31048573 | T | Educational attainment (years of education) | 30038396 | [0.0062-0.012] unit increase | 0.0091 | EFO_0004784 |
| ENSG00000165392 | rs62506104 | 8 | 31162492 | A | Educational attainment (years of education) | 30038396 | [0.0063-0.0133] unit decrease | 0.0098 | EFO_0004784 |
| ENSG00000165392 | rs1800389 | 8 | 31067041 | ? | Cardiovascular disease | 30595370 | EFO_0000319 |
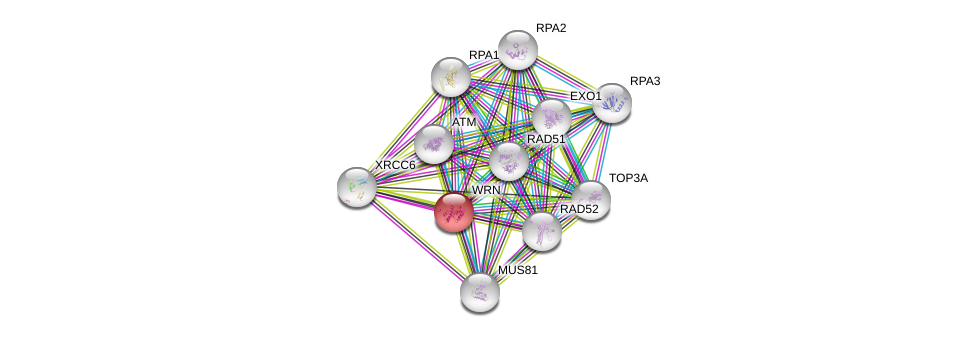
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000165392 | WRN | 81 | 69.805 | ENSAMEG00000000475 | - | 100 | 69.237 | Ailuropoda_melanoleuca |
| ENSG00000165392 | WRN | 92 | 55.049 | ENSAPLG00000001616 | WRN | 91 | 55.348 | Anas_platyrhynchos |
| ENSG00000165392 | WRN | 94 | 50.515 | ENSACAG00000003109 | WRN | 99 | 50.662 | Anolis_carolinensis |
| ENSG00000165392 | WRN | 100 | 85.625 | ENSANAG00000032806 | WRN | 99 | 89.774 | Aotus_nancymaae |
| ENSG00000165392 | WRN | 94 | 47.549 | ENSAMXG00000010066 | wrn | 89 | 46.896 | Astyanax_mexicanus |
| ENSG00000165392 | WRN | 100 | 72.292 | ENSBTAG00000021592 | WRN | 100 | 72.292 | Bos_taurus |
| ENSG00000165392 | WRN | 58 | 31.573 | WBGene00006944 | wrn-1 | 71 | 32.449 | Caenorhabditis_elegans |
| ENSG00000165392 | WRN | 100 | 88.494 | ENSCJAG00000012511 | WRN | 100 | 88.773 | Callithrix_jacchus |
| ENSG00000165392 | WRN | 100 | 72.207 | ENSCAFG00000006410 | WRN | 99 | 71.551 | Canis_familiaris |
| ENSG00000165392 | WRN | 100 | 73.943 | ENSCAFG00020001236 | WRN | 100 | 74.030 | Canis_lupus_dingo |
| ENSG00000165392 | WRN | 100 | 72.083 | ENSCHIG00000014991 | WRN | 100 | 72.083 | Capra_hircus |
| ENSG00000165392 | WRN | 99 | 78.457 | ENSTSYG00000012689 | WRN | 99 | 78.596 | Carlito_syrichta |
| ENSG00000165392 | WRN | 96 | 55.081 | ENSCAPG00000016803 | WRN | 98 | 70.882 | Cavia_aperea |
| ENSG00000165392 | WRN | 99 | 70.926 | ENSCPOG00000002943 | WRN | 99 | 70.791 | Cavia_porcellus |
| ENSG00000165392 | WRN | 100 | 87.997 | ENSCCAG00000023793 | WRN | 100 | 88.276 | Cebus_capucinus |
| ENSG00000165392 | WRN | 100 | 91.207 | ENSCATG00000040557 | WRN | 100 | 92.463 | Cercocebus_atys |
| ENSG00000165392 | WRN | 98 | 74.501 | ENSCLAG00000000817 | WRN | 100 | 71.218 | Chinchilla_lanigera |
| ENSG00000165392 | WRN | 96 | 93.827 | ENSCSAG00000015810 | WRN | 99 | 93.966 | Chlorocebus_sabaeus |
| ENSG00000165392 | WRN | 80 | 87.151 | ENSCHOG00000008966 | WRN | 81 | 87.151 | Choloepus_hoffmanni |
| ENSG00000165392 | WRN | 95 | 58.974 | ENSCPBG00000013421 | WRN | 90 | 59.151 | Chrysemys_picta_bellii |
| ENSG00000165392 | WRN | 64 | 37.846 | ENSCING00000002232 | - | 79 | 37.871 | Ciona_intestinalis |
| ENSG00000165392 | WRN | 100 | 93.510 | ENSCANG00000037137 | WRN | 100 | 93.022 | Colobus_angolensis_palliatus |
| ENSG00000165392 | WRN | 99 | 71.110 | ENSCGRG00001016491 | Wrn | 100 | 71.110 | Cricetulus_griseus_chok1gshd |
| ENSG00000165392 | WRN | 83 | 69.891 | ENSCGRG00000016821 | Wrn | 100 | 69.891 | Cricetulus_griseus_crigri |
| ENSG00000165392 | WRN | 94 | 47.237 | ENSDARG00000098211 | wrn | 89 | 46.951 | Danio_rerio |
| ENSG00000165392 | WRN | 95 | 71.418 | ENSDNOG00000008481 | WRN | 100 | 71.489 | Dasypus_novemcinctus |
| ENSG00000165392 | WRN | 99 | 71.399 | ENSDORG00000002696 | Wrn | 100 | 71.360 | Dipodomys_ordii |
| ENSG00000165392 | WRN | 77 | 80.676 | ENSETEG00000019854 | WRN | 79 | 80.676 | Echinops_telfairi |
| ENSG00000165392 | WRN | 98 | 70.526 | ENSEASG00005016813 | WRN | 98 | 80.986 | Equus_asinus_asinus |
| ENSG00000165392 | WRN | 100 | 71.968 | ENSECAG00000000110 | WRN | 100 | 72.035 | Equus_caballus |
| ENSG00000165392 | WRN | 85 | 64.789 | ENSEEUG00000004926 | WRN | 95 | 64.789 | Erinaceus_europaeus |
| ENSG00000165392 | WRN | 94 | 47.123 | ENSELUG00000017817 | wrn | 94 | 53.271 | Esox_lucius |
| ENSG00000165392 | WRN | 100 | 73.093 | ENSFCAG00000024759 | WRN | 100 | 72.935 | Felis_catus |
| ENSG00000165392 | WRN | 100 | 73.681 | ENSFDAG00000008637 | WRN | 100 | 73.342 | Fukomys_damarensis |
| ENSG00000165392 | WRN | 99 | 53.664 | ENSGALG00000040909 | WRN | 92 | 53.664 | Gallus_gallus |
| ENSG00000165392 | WRN | 99 | 55.608 | ENSGAGG00000022647 | WRN | 99 | 55.571 | Gopherus_agassizii |
| ENSG00000165392 | WRN | 100 | 96.718 | ENSGGOG00000022154 | WRN | 100 | 96.718 | Gorilla_gorilla |
| ENSG00000165392 | WRN | 99 | 73.989 | ENSHGLG00000003638 | WRN | 100 | 73.162 | Heterocephalus_glaber_female |
| ENSG00000165392 | WRN | 94 | 47.985 | ENSIPUG00000002090 | wrn | 92 | 47.935 | Ictalurus_punctatus |
| ENSG00000165392 | WRN | 100 | 74.797 | ENSSTOG00000013599 | WRN | 100 | 74.459 | Ictidomys_tridecemlineatus |
| ENSG00000165392 | WRN | 99 | 69.714 | ENSJJAG00000015655 | Wrn | 100 | 69.574 | Jaculus_jaculus |
| ENSG00000165392 | WRN | 97 | 49.929 | ENSLACG00000001569 | - | 93 | 50.071 | Latimeria_chalumnae |
| ENSG00000165392 | WRN | 95 | 51.055 | ENSLOCG00000010792 | wrn | 93 | 50.000 | Lepisosteus_oculatus |
| ENSG00000165392 | WRN | 99 | 74.654 | ENSLAFG00000004434 | WRN | 100 | 74.654 | Loxodonta_africana |
| ENSG00000165392 | WRN | 99 | 92.647 | ENSMFAG00000043428 | WRN | 99 | 93.557 | Macaca_fascicularis |
| ENSG00000165392 | WRN | 100 | 92.394 | ENSMMUG00000022655 | WRN | 100 | 93.301 | Macaca_mulatta |
| ENSG00000165392 | WRN | 100 | 93.719 | ENSMNEG00000032617 | WRN | 100 | 93.999 | Macaca_nemestrina |
| ENSG00000165392 | WRN | 100 | 92.184 | ENSMLEG00000038296 | WRN | 100 | 92.463 | Mandrillus_leucophaeus |
| ENSG00000165392 | WRN | 94 | 54.978 | ENSMGAG00000004960 | WRN | 88 | 54.792 | Meleagris_gallopavo |
| ENSG00000165392 | WRN | 99 | 71.179 | ENSMAUG00000016235 | Wrn | 100 | 71.179 | Mesocricetus_auratus |
| ENSG00000165392 | WRN | 99 | 74.478 | ENSMICG00000010630 | WRN | 100 | 74.983 | Microcebus_murinus |
| ENSG00000165392 | WRN | 99 | 70.133 | ENSMOCG00000013972 | Wrn | 100 | 70.063 | Microtus_ochrogaster |
| ENSG00000165392 | WRN | 100 | 55.163 | ENSMODG00000018997 | WRN | 100 | 55.532 | Monodelphis_domestica |
| ENSG00000165392 | WRN | 99 | 69.805 | MGP_CAROLIEiJ_G0030961 | Wrn | 100 | 69.526 | Mus_caroli |
| ENSG00000165392 | WRN | 99 | 70.014 | ENSMUSG00000031583 | Wrn | 100 | 69.735 | Mus_musculus |
| ENSG00000165392 | WRN | 99 | 69.526 | MGP_PahariEiJ_G0021430 | Wrn | 100 | 69.247 | Mus_pahari |
| ENSG00000165392 | WRN | 99 | 70.223 | MGP_SPRETEiJ_G0032079 | Wrn | 100 | 69.944 | Mus_spretus |
| ENSG00000165392 | WRN | 100 | 73.162 | ENSMPUG00000010201 | WRN | 100 | 73.162 | Mustela_putorius_furo |
| ENSG00000165392 | WRN | 99 | 75.785 | ENSMLUG00000005863 | WRN | 99 | 75.785 | Myotis_lucifugus |
| ENSG00000165392 | WRN | 99 | 72.176 | ENSNGAG00000018413 | Wrn | 99 | 72.176 | Nannospalax_galili |
| ENSG00000165392 | WRN | 100 | 96.371 | ENSNLEG00000012006 | WRN | 100 | 96.371 | Nomascus_leucogenys |
| ENSG00000165392 | WRN | 73 | 63.755 | ENSMEUG00000008271 | - | 76 | 63.478 | Notamacropus_eugenii |
| ENSG00000165392 | WRN | 89 | 63.994 | ENSOPRG00000001480 | WRN | 88 | 63.893 | Ochotona_princeps |
| ENSG00000165392 | WRN | 99 | 73.449 | ENSODEG00000004076 | WRN | 100 | 73.101 | Octodon_degus |
| ENSG00000165392 | WRN | 89 | 61.479 | ENSOANG00000005952 | - | 93 | 61.479 | Ornithorhynchus_anatinus |
| ENSG00000165392 | WRN | 100 | 75.554 | ENSOCUG00000012620 | WRN | 90 | 75.798 | Oryctolagus_cuniculus |
| ENSG00000165392 | WRN | 97 | 73.714 | ENSOGAG00000009600 | WRN | 100 | 73.714 | Otolemur_garnettii |
| ENSG00000165392 | WRN | 100 | 72.222 | ENSOARG00000000163 | WRN | 100 | 72.222 | Ovis_aries |
| ENSG00000165392 | WRN | 100 | 96.718 | ENSPPAG00000036871 | WRN | 100 | 96.718 | Pan_paniscus |
| ENSG00000165392 | WRN | 100 | 74.254 | ENSPPRG00000010928 | WRN | 100 | 74.046 | Panthera_pardus |
| ENSG00000165392 | WRN | 100 | 74.046 | ENSPTIG00000020609 | WRN | 100 | 73.838 | Panthera_tigris_altaica |
| ENSG00000165392 | WRN | 100 | 98.394 | ENSPTRG00000020145 | WRN | 100 | 98.394 | Pan_troglodytes |
| ENSG00000165392 | WRN | 100 | 92.817 | ENSPANG00000018594 | WRN | 100 | 93.445 | Papio_anubis |
| ENSG00000165392 | WRN | 70 | 67.262 | ENSPKIG00000025497 | wrn | 93 | 60.039 | Paramormyrops_kingsleyae |
| ENSG00000165392 | WRN | 94 | 57.768 | ENSPSIG00000013411 | WRN | 89 | 58.284 | Pelodiscus_sinensis |
| ENSG00000165392 | WRN | 99 | 69.784 | ENSPEMG00000020615 | Wrn | 99 | 70.118 | Peromyscus_maniculatus_bairdii |
| ENSG00000165392 | WRN | 97 | 55.713 | ENSPCIG00000020146 | WRN | 99 | 56.407 | Phascolarctos_cinereus |
| ENSG00000165392 | WRN | 100 | 96.439 | ENSPPYG00000018494 | WRN | 100 | 96.439 | Pongo_abelii |
| ENSG00000165392 | WRN | 88 | 70.690 | ENSPCAG00000013763 | WRN | 88 | 70.690 | Procavia_capensis |
| ENSG00000165392 | WRN | 79 | 66.824 | ENSPCOG00000012938 | WRN | 98 | 66.667 | Propithecus_coquereli |
| ENSG00000165392 | WRN | 96 | 74.257 | ENSPVAG00000012909 | WRN | 96 | 74.257 | Pteropus_vampyrus |
| ENSG00000165392 | WRN | 94 | 46.998 | ENSPNAG00000021848 | wrn | 88 | 46.998 | Pygocentrus_nattereri |
| ENSG00000165392 | WRN | 99 | 69.225 | ENSRNOG00000015440 | Wrn | 100 | 69.156 | Rattus_norvegicus |
| ENSG00000165392 | WRN | 100 | 90.021 | ENSRBIG00000031825 | WRN | 100 | 90.902 | Rhinopithecus_bieti |
| ENSG00000165392 | WRN | 100 | 93.231 | ENSRROG00000045381 | WRN | 100 | 93.440 | Rhinopithecus_roxellana |
| ENSG00000165392 | WRN | 100 | 88.904 | ENSSBOG00000027238 | WRN | 100 | 88.555 | Saimiri_boliviensis_boliviensis |
| ENSG00000165392 | WRN | 99 | 57.626 | ENSSHAG00000005577 | WRN | 99 | 57.833 | Sarcophilus_harrisii |
| ENSG00000165392 | WRN | 95 | 48.148 | ENSSFOG00015006123 | wrn | 95 | 47.962 | Scleropages_formosus |
| ENSG00000165392 | WRN | 74 | 65.368 | ENSSARG00000006157 | WRN | 87 | 65.368 | Sorex_araneus |
| ENSG00000165392 | WRN | 97 | 76.450 | ENSSSCG00000015840 | WRN | 100 | 76.450 | Sus_scrofa |
| ENSG00000165392 | WRN | 72 | 57.336 | ENSTGUG00000001765 | WRN | 100 | 57.336 | Taeniopygia_guttata |
| ENSG00000165392 | WRN | 78 | 62.372 | ENSTBEG00000012702 | - | 78 | 62.030 | Tupaia_belangeri |
| ENSG00000165392 | WRN | 100 | 70.744 | ENSTTRG00000012302 | WRN | 100 | 70.151 | Tursiops_truncatus |
| ENSG00000165392 | WRN | 100 | 71.221 | ENSUAMG00000021324 | WRN | 100 | 71.498 | Ursus_americanus |
| ENSG00000165392 | WRN | 86 | 71.637 | ENSUMAG00000007159 | WRN | 97 | 71.961 | Ursus_maritimus |
| ENSG00000165392 | WRN | 99 | 71.329 | ENSVPAG00000011004 | WRN | 99 | 71.329 | Vicugna_pacos |
| ENSG00000165392 | WRN | 100 | 72.005 | ENSVVUG00000019867 | WRN | 100 | 72.091 | Vulpes_vulpes |
| ENSG00000165392 | WRN | 98 | 52.572 | ENSXETG00000018302 | wrn | 99 | 53.834 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | 21873635. | IBA | Function |
| GO:0000287 | magnesium ion binding | 16622405. | IDA | Function |
| GO:0000400 | four-way junction DNA binding | 21873635. | IBA | Function |
| GO:0000400 | four-way junction DNA binding | 11735402. | IDA | Function |
| GO:0000403 | Y-form DNA binding | 21873635. | IBA | Function |
| GO:0000403 | Y-form DNA binding | 11735402.17715146.26420422. | IDA | Function |
| GO:0000405 | bubble DNA binding | 21873635. | IBA | Function |
| GO:0000405 | bubble DNA binding | 11433031.11735402. | IDA | Function |
| GO:0000723 | telomere maintenance | 18212065. | IMP | Process |
| GO:0000723 | telomere maintenance | 25801465. | TAS | Process |
| GO:0000724 | double-strand break repair via homologous recombination | 21873635. | IBA | Process |
| GO:0000731 | DNA synthesis involved in DNA repair | 21873635. | IBA | Process |
| GO:0000731 | DNA synthesis involved in DNA repair | 17563354. | IDA | Process |
| GO:0000781 | chromosome, telomeric region | 21873635. | IBA | Component |
| GO:0000781 | chromosome, telomeric region | 15200954.26420422. | IDA | Component |
| GO:0001302 | replicative cell aging | 21873635. | IBA | Process |
| GO:0003677 | DNA binding | 21873635. | IBA | Function |
| GO:0003677 | DNA binding | 9288107. | IDA | Function |
| GO:0003678 | DNA helicase activity | 12944467.19734539.26420422. | IDA | Function |
| GO:0003678 | DNA helicase activity | 10871376. | IMP | Function |
| GO:0003682 | chromatin binding | 21873635. | IBA | Function |
| GO:0004003 | ATP-dependent DNA helicase activity | 9288107.15200954. | IDA | Function |
| GO:0004386 | helicase activity | 12181313. | IDA | Function |
| GO:0004527 | exonuclease activity | 12944467. | IDA | Function |
| GO:0004527 | exonuclease activity | 15200954. | IMP | Function |
| GO:0005515 | protein binding | 10373438.10783163.11328876.11427532.11598021.11919194.12181313.12750383.14596914.14657243.14688284.14734561.15735006.15965237.16030011.17498979.17611195.17715146.17961633.18203716.19343720.21639834.24126761.25931448.26496610. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | 11420665.12944467.17498979.21639834.25801465. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005657 | replication fork | 21873635. | IBA | Component |
| GO:0005657 | replication fork | - | ISS | Component |
| GO:0005662 | DNA replication factor A complex | 21873635. | IBA | Component |
| GO:0005694 | chromosome | 21873635. | IBA | Component |
| GO:0005730 | nucleolus | 9618508.11420665.12181313.12944467.15200954.17498979.21639834.25801465. | IDA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005813 | centrosome | 21873635. | IBA | Component |
| GO:0005813 | centrosome | 17498979. | IDA | Component |
| GO:0006259 | DNA metabolic process | 16622405. | IDA | Process |
| GO:0006260 | DNA replication | 12882351. | IMP | Process |
| GO:0006260 | DNA replication | - | ISS | Process |
| GO:0006260 | DNA replication | - | TAS | Process |
| GO:0006281 | DNA repair | 21873635. | IBA | Process |
| GO:0006284 | base-excision repair | 21873635. | IBA | Process |
| GO:0006284 | base-excision repair | 17611195. | IDA | Process |
| GO:0006302 | double-strand break repair | 21639834. | IMP | Process |
| GO:0006310 | DNA recombination | 21873635. | IBA | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 18203716. | IDA | Process |
| GO:0006979 | response to oxidative stress | 21873635. | IBA | Process |
| GO:0006979 | response to oxidative stress | 17611195. | IDA | Process |
| GO:0007420 | brain development | - | IEA | Process |
| GO:0007568 | aging | 9288107. | NAS | Process |
| GO:0007569 | cell aging | 21873635. | IBA | Process |
| GO:0007569 | cell aging | 18212065. | IMP | Process |
| GO:0008340 | determination of adult lifespan | 21873635. | IBA | Process |
| GO:0008408 | 3'-5' exonuclease activity | 21873635. | IBA | Function |
| GO:0008408 | 3'-5' exonuclease activity | 10783163.10871373.12181313.16622405. | IDA | Function |
| GO:0009267 | cellular response to starvation | 21873635. | IBA | Process |
| GO:0009267 | cellular response to starvation | 11420665. | IDA | Process |
| GO:0009378 | four-way junction helicase activity | 21873635. | IBA | Function |
| GO:0009378 | four-way junction helicase activity | 11433031.11735402. | IDA | Function |
| GO:0010225 | response to UV-C | 21873635. | IBA | Process |
| GO:0010225 | response to UV-C | 17563354. | IDA | Process |
| GO:0010259 | multicellular organism aging | 21873635. | IBA | Process |
| GO:0010259 | multicellular organism aging | 16673358. | IMP | Process |
| GO:0016607 | nuclear speck | - | IDA | Component |
| GO:0016887 | ATPase activity | 10373438.10871376. | IDA | Function |
| GO:0030145 | manganese ion binding | 21873635. | IBA | Function |
| GO:0030145 | manganese ion binding | 16622405. | IDA | Function |
| GO:0031297 | replication fork processing | 21873635. | IBA | Process |
| GO:0031297 | replication fork processing | 17115688. | IDA | Process |
| GO:0031297 | replication fork processing | 12882351. | IMP | Process |
| GO:0032405 | MutLalpha complex binding | 17715146. | IDA | Function |
| GO:0032508 | DNA duplex unwinding | 21873635. | IBA | Process |
| GO:0032508 | DNA duplex unwinding | 11735402.26420422. | IDA | Process |
| GO:0035861 | site of double-strand break | 21873635. | IBA | Component |
| GO:0040009 | regulation of growth rate | - | IEA | Process |
| GO:0042803 | protein homodimerization activity | 21873635. | IBA | Function |
| GO:0042803 | protein homodimerization activity | 10783163. | IDA | Function |
| GO:0042981 | regulation of apoptotic process | 21873635. | IBA | Process |
| GO:0042981 | regulation of apoptotic process | 9681877. | IGI | Process |
| GO:0043005 | neuron projection | - | IEA | Component |
| GO:0043138 | 3'-5' DNA helicase activity | 17715146. | IDA | Function |
| GO:0043140 | ATP-dependent 3'-5' DNA helicase activity | 21873635. | IBA | Function |
| GO:0044806 | G-quadruplex DNA unwinding | 21873635. | IBA | Process |
| GO:0044806 | G-quadruplex DNA unwinding | 11735402. | IDA | Process |
| GO:0044806 | G-quadruplex DNA unwinding | - | ISS | Process |
| GO:0044877 | protein-containing complex binding | 21873635. | IBA | Function |
| GO:0044877 | protein-containing complex binding | 10783163. | IDA | Function |
| GO:0051345 | positive regulation of hydrolase activity | 21873635. | IBA | Process |
| GO:0051345 | positive regulation of hydrolase activity | 17611195. | IDA | Process |
| GO:0051880 | G-quadruplex DNA binding | 11433031.11735402. | IDA | Function |
| GO:0061749 | forked DNA-dependent helicase activity | 21873635. | IBA | Function |
| GO:0061749 | forked DNA-dependent helicase activity | 11735402. | IDA | Function |
| GO:0061820 | telomeric D-loop disassembly | 21873635. | IBA | Process |
| GO:0061820 | telomeric D-loop disassembly | 15200954.19734539.26420422. | IDA | Process |
| GO:0061820 | telomeric D-loop disassembly | 22039056. | IGI | Process |
| GO:0061820 | telomeric D-loop disassembly | 22039056. | TAS | Process |
| GO:0061821 | telomeric D-loop binding | 21873635. | IBA | Function |
| GO:0061821 | telomeric D-loop binding | 19734539.26420422. | IDA | Function |
| GO:0061849 | telomeric G-quadruplex DNA binding | 19734539. | IC | Function |
| GO:0070337 | 3'-flap-structured DNA binding | 21873635. | IBA | Function |
| GO:0070337 | 3'-flap-structured DNA binding | 26420422. | IDA | Function |
| GO:0071480 | cellular response to gamma radiation | 21873635. | IBA | Process |
| GO:0071480 | cellular response to gamma radiation | 21639834. | IDA | Process |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | - | IEA | Process |
| GO:0090656 | t-circle formation | 27918544. | TAS | Process |
| GO:0098530 | positive regulation of strand invasion | 21873635. | IBA | Process |
| GO:0098530 | positive regulation of strand invasion | 26420422. | IDA | Process |
| GO:1901796 | regulation of signal transduction by p53 class mediator | - | TAS | Process |
| GO:1902570 | protein localization to nucleolus | 11420665. | IDA | Process |
| GO:1905773 | 8-hydroxy-2'-deoxyguanosine DNA binding | 21873635. | IBA | Function |
| GO:1905773 | 8-hydroxy-2'-deoxyguanosine DNA binding | 19734539. | IDA | Function |