
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ESCA | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| SKCM | ||
| BLCA | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| LUAD |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 21282044 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000537508 | SLC3A2-209 | 583 | - | - | - (aa) | - | - |
| ENST00000457660 | SLC3A2-205 | 873 | - | - | - (aa) | - | - |
| ENST00000542793 | SLC3A2-219 | 539 | - | - | - (aa) | - | - |
| ENST00000535296 | SLC3A2-206 | 2084 | - | ENSP00000444236 | 599 (aa) | - | F5GZS6 |
| ENST00000536981 | SLC3A2-208 | 1486 | - | ENSP00000444439 | 175 (aa) | - | F5GZI0 |
| ENST00000377891 | SLC3A2-204 | 2222 | - | ENSP00000367123 | 631 (aa) | - | J3KPF3 |
| ENST00000377890 | SLC3A2-203 | 2161 | - | ENSP00000367122 | 630 (aa) | - | P08195 |
| ENST00000539458 | SLC3A2-213 | 565 | - | ENSP00000438032 | 145 (aa) | - | F5H0E2 |
| ENST00000546253 | SLC3A2-222 | 582 | - | - | - (aa) | - | - |
| ENST00000537839 | SLC3A2-210 | 584 | - | - | - (aa) | - | - |
| ENST00000538682 | SLC3A2-212 | 1368 | - | - | - (aa) | - | - |
| ENST00000535768 | SLC3A2-207 | 597 | - | - | - (aa) | - | - |
| ENST00000544377 | SLC3A2-221 | 589 | - | ENSP00000442135 | 135 (aa) | - | F5H867 |
| ENST00000539891 | SLC3A2-215 | 1300 | - | ENSP00000438353 | 67 (aa) | - | F5H056 |
| ENST00000542922 | SLC3A2-220 | 2174 | - | - | - (aa) | - | - |
| ENST00000546312 | SLC3A2-223 | 564 | - | - | - (aa) | - | - |
| ENST00000541372 | SLC3A2-216 | 269 | - | ENSP00000444250 | 47 (aa) | - | F5GZR9 |
| ENST00000539507 | SLC3A2-214 | 534 | - | ENSP00000440819 | 122 (aa) | - | H0YFX4 |
| ENST00000377889 | SLC3A2-202 | 1993 | - | ENSP00000367121 | 568 (aa) | - | P08195 |
| ENST00000541425 | SLC3A2-217 | 595 | - | ENSP00000445858 | 24 (aa) | - | H0YH36 |
| ENST00000541649 | SLC3A2-218 | 515 | - | - | - (aa) | - | - |
| ENST00000338663 | SLC3A2-201 | 1904 | - | ENSP00000340815 | 529 (aa) | - | P08195 |
| ENST00000538084 | SLC3A2-211 | 741 | - | ENSP00000440001 | 247 (aa) | - | H0YFS2 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000168003 | Myocardial Infarction | 5.8015742E-004 | - |
| ENSG00000168003 | Myocardial Infarction | 2.6274260E-004 | - |
| ENSG00000168003 | Heart Failure | 6.1115535E-004 | - |
| ENSG00000168003 | Platelet Function Tests | 7.4600000E-006 | - |
| ENSG00000168003 | Platelet Function Tests | 2.5900000E-006 | - |
| ENSG00000168003 | Platelet Function Tests | 1.1263300E-005 | - |
| ENSG00000168003 | Platelet Function Tests | 1.7172800E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000168003 | rs144453006 | 11 | 62887212 | G | Iron status biomarkers (transferrin saturation) | 28334935 | [0.57-1.32] unit decrease | 0.946 | EFO_0006333 |
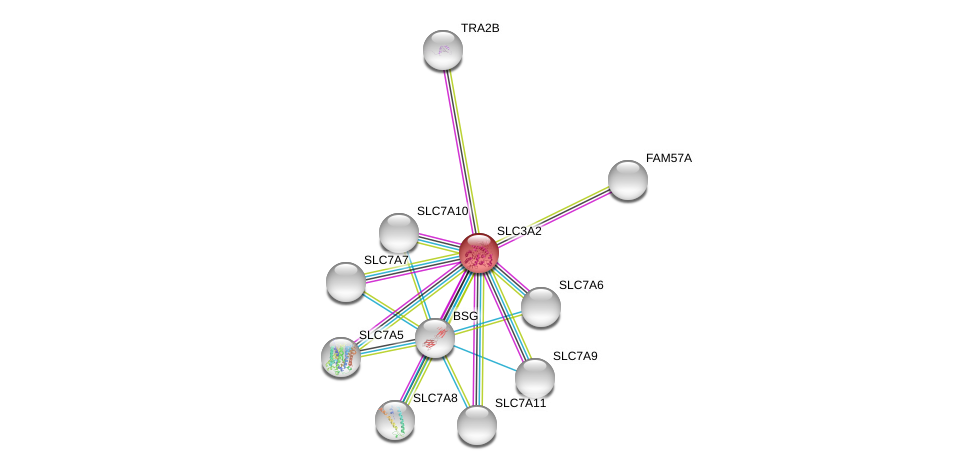
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000168003 | SLC3A2 | 100 | 86.667 | ENSMUSG00000010095 | Slc3a2 | 100 | 87.826 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0003725 | double-stranded RNA binding | 21266579. | IDA | Function |
| GO:0003824 | catalytic activity | - | IEA | Function |
| GO:0005432 | calcium:sodium antiporter activity | 10673541. | TAS | Function |
| GO:0005515 | protein binding | 10506149.10631289.12270127.20374249.25998567.28298427. | IPI | Function |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005886 | plasma membrane | 12270127.25998567. | IDA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005975 | carbohydrate metabolic process | - | IEA | Process |
| GO:0006816 | calcium ion transport | 3036867. | NAS | Process |
| GO:0006865 | amino acid transport | 10673541. | TAS | Process |
| GO:0009986 | cell surface | 19581412. | HDA | Component |
| GO:0009986 | cell surface | 25063885. | IDA | Component |
| GO:0015175 | neutral amino acid transmembrane transporter activity | - | ISS | Function |
| GO:0015827 | tryptophan transport | - | ISS | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016021 | integral component of membrane | 3476959. | NAS | Component |
| GO:0016324 | apical plasma membrane | - | IEA | Component |
| GO:0035725 | sodium ion transmembrane transport | - | IEA | Process |
| GO:0042470 | melanosome | - | IEA | Component |
| GO:0043330 | response to exogenous dsRNA | 21266579. | IMP | Process |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0050900 | leukocyte migration | - | TAS | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.23533145. | HDA | Component |
| GO:1902475 | L-alpha-amino acid transmembrane transport | - | TAS | Process |
| GO:1903801 | L-leucine import across plasma membrane | - | ISS | Process |