
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 18307534 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| KIRC | |||
| READ | |||
| TGCT | |||
| THCA | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| SKCM | ||
| BRCA |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000343893 | S1-like | PF14444.6 | 1.5e-19 | 1 | 2 |
| ENSP00000343893 | S1-like | PF14444.6 | 1.5e-19 | 2 | 2 |
| ENSP00000276241 | S1-like | PF14444.6 | 2.2e-19 | 1 | 2 |
| ENSP00000276241 | S1-like | PF14444.6 | 2.2e-19 | 2 | 2 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000276241 | CT55-201 | 1445 | XM_005262427 | ENSP00000276241 | 264 (aa) | XP_005262484 | Q8WUE5 |
| ENST00000344129 | CT55-202 | 1728 | - | ENSP00000343893 | 242 (aa) | - | Q8WUE5 |
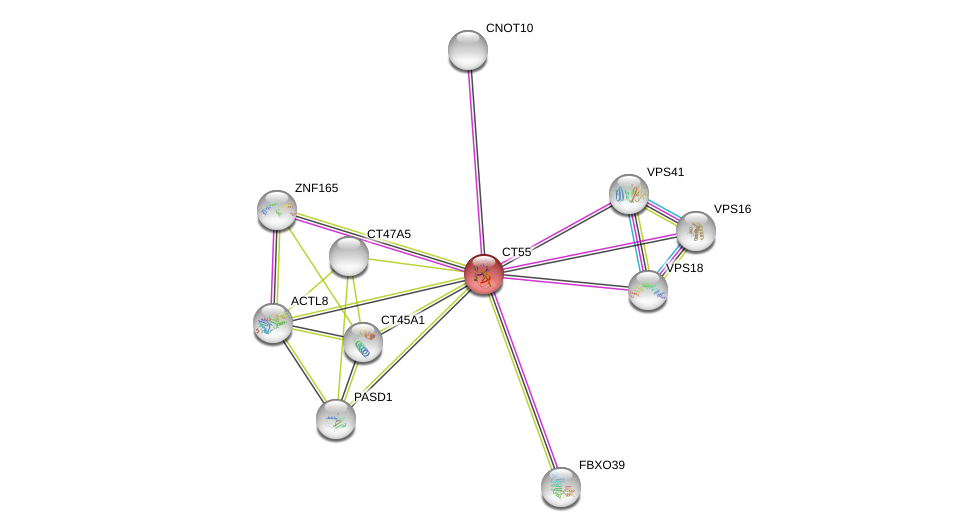
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000169551 | CT55 | 95 | 69.323 | ENSANAG00000025850 | CT55 | 96 | 67.323 | Aotus_nancymaae |
| ENSG00000169551 | CT55 | 100 | 71.074 | ENSCJAG00000020403 | CT55 | 95 | 69.444 | Callithrix_jacchus |
| ENSG00000169551 | CT55 | 94 | 73.387 | ENSCCAG00000033288 | CT55 | 96 | 72.441 | Cebus_capucinus |
| ENSG00000169551 | CT55 | 95 | 83.200 | ENSCATG00000032930 | CT55 | 96 | 82.677 | Cercocebus_atys |
| ENSG00000169551 | CT55 | 96 | 89.370 | ENSCSAG00000007710 | - | 96 | 89.370 | Chlorocebus_sabaeus |
| ENSG00000169551 | CT55 | 94 | 79.151 | ENSCSAG00000007720 | - | 96 | 78.409 | Chlorocebus_sabaeus |
| ENSG00000169551 | CT55 | 100 | 82.197 | ENSCANG00000038605 | CT55 | 96 | 81.890 | Colobus_angolensis_palliatus |
| ENSG00000169551 | CT55 | 58 | 97.163 | ENSGGOG00000028404 | - | 90 | 97.163 | Gorilla_gorilla |
| ENSG00000169551 | CT55 | 95 | 85.600 | ENSMFAG00000031424 | CT55 | 96 | 85.039 | Macaca_fascicularis |
| ENSG00000169551 | CT55 | 95 | 84.064 | ENSMMUG00000018715 | CT55 | 96 | 83.529 | Macaca_mulatta |
| ENSG00000169551 | CT55 | 95 | 85.600 | ENSMNEG00000037841 | CT55 | 96 | 85.039 | Macaca_nemestrina |
| ENSG00000169551 | CT55 | 95 | 82.800 | ENSMLEG00000028336 | CT55 | 96 | 82.283 | Mandrillus_leucophaeus |
| ENSG00000169551 | CT55 | 100 | 64.198 | ENSMICG00000047795 | CT55 | 95 | 60.558 | Microcebus_murinus |
| ENSG00000169551 | CT55 | 96 | 93.701 | ENSNLEG00000029349 | CT55 | 96 | 93.701 | Nomascus_leucogenys |
| ENSG00000169551 | CT55 | 99 | 59.167 | ENSOGAG00000007138 | CT55 | 96 | 59.592 | Otolemur_garnettii |
| ENSG00000169551 | CT55 | 74 | 97.207 | ENSPPAG00000001184 | - | 100 | 97.207 | Pan_paniscus |
| ENSG00000169551 | CT55 | 65 | 89.474 | ENSPTRG00000022296 | CT55 | 100 | 89.474 | Pan_troglodytes |
| ENSG00000169551 | CT55 | 100 | 97.934 | ENSPTRG00000050752 | CT55 | 100 | 97.727 | Pan_troglodytes |
| ENSG00000169551 | CT55 | 94 | 84.141 | ENSPANG00000012503 | CT55 | 95 | 83.550 | Papio_anubis |
| ENSG00000169551 | CT55 | 96 | 92.520 | ENSPPYG00000020744 | CT55 | 96 | 92.520 | Pongo_abelii |
| ENSG00000169551 | CT55 | 100 | 63.223 | ENSPCOG00000015916 | CT55 | 96 | 61.811 | Propithecus_coquereli |
| ENSG00000169551 | CT55 | 99 | 44.583 | ENSRNOG00000061094 | LOC100360296 | 87 | 46.635 | Rattus_norvegicus |
| ENSG00000169551 | CT55 | 99 | 49.793 | ENSRBIG00000032050 | - | 95 | 52.381 | Rhinopithecus_bieti |
| ENSG00000169551 | CT55 | 96 | 89.224 | ENSRBIG00000036226 | - | 96 | 89.224 | Rhinopithecus_bieti |
| ENSG00000169551 | CT55 | 100 | 82.197 | ENSRROG00000043598 | CT55 | 96 | 82.283 | Rhinopithecus_roxellana |
| ENSG00000169551 | CT55 | 86 | 63.462 | ENSTBEG00000009423 | - | 99 | 60.699 | Tupaia_belangeri |
| ENSG00000169551 | CT55 | 95 | 56.332 | ENSTBEG00000013745 | - | 99 | 56.332 | Tupaia_belangeri |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 25416956. | IPI | Function |