
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| DLBC | |||
| THCA | |||
| UCEC | |||
| UCS | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SKCM | ||
| PAAD | ||
| READ | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LGG | ||
| LUAD | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000449889 | JUP-208 | 1069 | - | ENSP00000389886 | 297 (aa) | - | C9JKY1 |
| ENST00000437187 | JUP-206 | 1029 | - | ENSP00000394146 | 295 (aa) | - | C9JK18 |
| ENST00000585793 | JUP-210 | 598 | - | - | - (aa) | - | - |
| ENST00000437369 | JUP-207 | 586 | - | ENSP00000409948 | 86 (aa) | - | C9JPI2 |
| ENST00000465293 | JUP-209 | 432 | - | ENSP00000467065 | 2 (aa) | - | UPI000011EBE6 |
| ENST00000393930 | JUP-202 | 3298 | - | ENSP00000377507 | 745 (aa) | - | P14923 |
| ENST00000393931 | JUP-203 | 3497 | - | ENSP00000377508 | 745 (aa) | - | P14923 |
| ENST00000424457 | JUP-205 | 949 | - | ENSP00000401034 | 235 (aa) | - | C9JTX4 |
| ENST00000310706 | JUP-201 | 3200 | XM_006721874 | ENSP00000311113 | 745 (aa) | XP_006721937 | P14923 |
| ENST00000420370 | JUP-204 | 915 | - | ENSP00000411449 | 276 (aa) | - | C9J826 |
| ENST00000591690 | JUP-212 | 583 | - | ENSP00000468347 | 104 (aa) | - | K7ERP3 |
| ENST00000589036 | JUP-211 | 566 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000173801 | rs143043662 | 17 | 41757519 | C | Heel bone mineral density | 28869591 | [0.059-0.116] unit decrease | 0.0876303 | EFO_0009270 |
| ENSG00000173801 | rs143043662 | 17 | 41757519 | C | Heel bone mineral density | 28869591 | [0.048-0.126] unit decrease | 0.0872999 | EFO_0009270 |
| ENSG00000173801 | rs143043662 | 17 | 41757519 | ? | Heel bone mineral density | 30048462 | [0.059-0.097] unit decrease | 0.0780376 | EFO_0009270 |
| ENSG00000173801 | rs7213095 | 17 | 41784516 | ? | Heel bone mineral density | 30048462 | [0.0082-0.017] unit decrease | 0.0125896 | EFO_0009270 |
| ENSG00000173801 | rs143043662 | 17 | 41757519 | C | Heel bone mineral density | 30598549 | [0.052-0.086] unit decrease | 0.0687204 | EFO_0009270 |
| ENSG00000173801 | rs41283425 | 17 | 41769461 | ? | Balding type 1 | 30595370 | EFO_0007825 | ||
| ENSG00000173801 | rs41283425 | 17 | 41769461 | T | Male-pattern baldness | 30573740 | [0.042-0.07] unit increase | 0.0559405 | EFO_0007825 |
| ENSG00000173801 | rs143043662 | 17 | 41757519 | ? | Heel bone mineral density | 30595370 | EFO_0009270 |
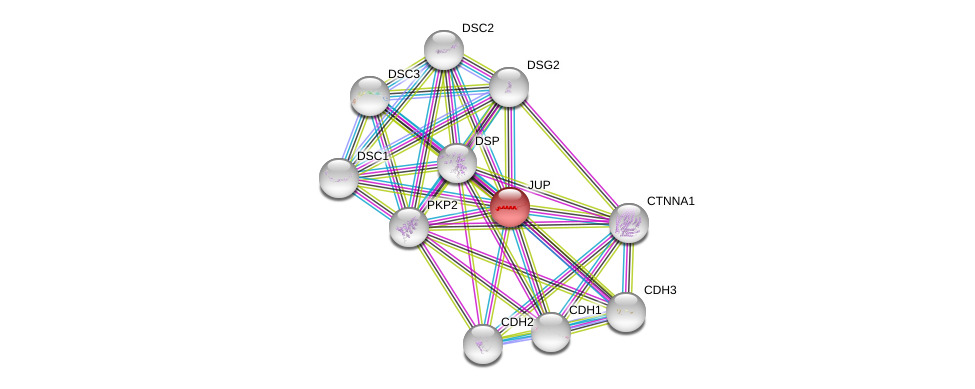
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000173801 | JUP | 100 | 98.389 | ENSMUSG00000001552 | Jup | 100 | 98.389 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001533 | cornified envelope | - | TAS | Component |
| GO:0001954 | positive regulation of cell-matrix adhesion | 28115160. | IGI | Process |
| GO:0002159 | desmosome assembly | 22889254. | IDA | Process |
| GO:0002159 | desmosome assembly | 20859650. | IMP | Process |
| GO:0003713 | transcription coactivator activity | 21873635. | IBA | Function |
| GO:0003713 | transcription coactivator activity | 14661054. | IDA | Function |
| GO:0005198 | structural molecule activity | 22889254. | NAS | Function |
| GO:0005199 | structural constituent of cell wall | 7650039. | IC | Function |
| GO:0005515 | protein binding | 7650039.7890674.7983064.10801826.12370829.14661054.16189514.16917092.17098867.17510365.19060904.20859650.20936779.24965446.25241761.25416956.26496610.26871637. | IPI | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005634 | nucleus | 14661054. | IMP | Component |
| GO:0005737 | cytoplasm | 14661054. | IMP | Component |
| GO:0005829 | cytosol | - | ISS | Component |
| GO:0005856 | cytoskeleton | - | ISS | Component |
| GO:0005882 | intermediate filament | - | IEA | Component |
| GO:0005886 | plasma membrane | 14661054.20859650. | IDA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005911 | cell-cell junction | 1639850.7650039.7983064.10460003.14661054. | IDA | Component |
| GO:0005913 | cell-cell adherens junction | 21873635. | IBA | Component |
| GO:0005913 | cell-cell adherens junction | 10403777. | IDA | Component |
| GO:0005915 | zonula adherens | - | ISS | Component |
| GO:0005916 | fascia adherens | - | IEA | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0007016 | cytoskeletal anchoring at plasma membrane | 22889254. | NAS | Process |
| GO:0009898 | cytoplasmic side of plasma membrane | - | ISS | Component |
| GO:0014704 | intercalated disc | 22781308.22889254.23381804. | IDA | Component |
| GO:0015629 | actin cytoskeleton | - | IEA | Component |
| GO:0016327 | apicolateral plasma membrane | - | IEA | Component |
| GO:0016328 | lateral plasma membrane | - | IEA | Component |
| GO:0016342 | catenin complex | 21873635. | IBA | Component |
| GO:0016342 | catenin complex | 7650039. | IDA | Component |
| GO:0016477 | cell migration | 18937352. | IMP | Process |
| GO:0019901 | protein kinase binding | - | IEA | Function |
| GO:0019903 | protein phosphatase binding | 21873635. | IBA | Function |
| GO:0019903 | protein phosphatase binding | 8663237. | IPI | Function |
| GO:0030018 | Z disc | - | IEA | Component |
| GO:0030056 | hemidesmosome | - | ISS | Component |
| GO:0030057 | desmosome | 1639850. | IDA | Component |
| GO:0031424 | keratinization | - | TAS | Process |
| GO:0032993 | protein-DNA complex | 14661054. | IDA | Component |
| GO:0034332 | adherens junction organization | - | TAS | Process |
| GO:0035257 | nuclear hormone receptor binding | 21873635. | IBA | Function |
| GO:0035580 | specific granule lumen | - | TAS | Component |
| GO:0042127 | regulation of cell proliferation | 17924338. | IDA | Process |
| GO:0042307 | positive regulation of protein import into nucleus | 10825188.14661054. | IDA | Process |
| GO:0042803 | protein homodimerization activity | - | ISS | Function |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0043537 | negative regulation of blood vessel endothelial cell migration | 28115160. | IGI | Process |
| GO:0045294 | alpha-catenin binding | 21873635. | IBA | Function |
| GO:0045294 | alpha-catenin binding | 7650039.7890674.23136403. | IPI | Function |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0045296 | cadherin binding | 21873635. | IBA | Function |
| GO:0045296 | cadherin binding | 1639850.7650039. | IPI | Function |
| GO:0045766 | positive regulation of angiogenesis | 28115160. | IGI | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0050839 | cell adhesion molecule binding | 17559062. | IPI | Function |
| GO:0050982 | detection of mechanical stimulus | 18937352. | IDA | Process |
| GO:0051091 | positive regulation of DNA-binding transcription factor activity | 14661054. | IDA | Process |
| GO:0051291 | protein heterooligomerization | - | IEA | Process |
| GO:0070062 | extracellular exosome | 19056867. | HDA | Component |
| GO:0070268 | cornification | - | TAS | Process |
| GO:0071603 | endothelial cell-cell adhesion | - | ISS | Process |
| GO:0071665 | gamma-catenin-TCF7L2 complex | 14661054. | IDA | Component |
| GO:0071681 | cellular response to indole-3-methanol | 10868478. | IDA | Process |
| GO:0072659 | protein localization to plasma membrane | 20859650. | IMP | Process |
| GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 17924338. | IMP | Process |
| GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication | 17924338. | IC | Function |
| GO:0086091 | regulation of heart rate by cardiac conduction | 17924338. | IMP | Process |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | 14661054. | IC | Process |
| GO:0098609 | cell-cell adhesion | 18937352. | IDA | Process |
| GO:0098609 | cell-cell adhesion | 20859650. | IMP | Process |
| GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 17924338. | IMP | Process |
| GO:1904813 | ficolin-1-rich granule lumen | - | TAS | Component |