
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| HNSC | |||
| LUAD | |||
| LUSC | |||
| PAAD | |||
| PRAD | |||
| SKCM |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| BRCA | ||
| CESC | ||
| UCEC | ||
| LUAD | ||
| UVM |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 11677214 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000313432 | ATP2A2-202 | 5575 | - | - | - (aa) | - | - |
| ENST00000547792 | ATP2A2-206 | 541 | - | - | - (aa) | - | - |
| ENST00000547050 | ATP2A2-205 | 1176 | - | - | - (aa) | - | - |
| ENST00000553144 | ATP2A2-212 | 264 | - | ENSP00000450407 | 46 (aa) | - | A0A0C4DH86 |
| ENST00000539276 | ATP2A2-204 | 4094 | XM_005253888 | ENSP00000440045 | 1042 (aa) | XP_005253945 | P16615 |
| ENST00000550262 | ATP2A2-210 | 3670 | - | - | - (aa) | - | - |
| ENST00000308664 | ATP2A2-201 | 4453 | - | ENSP00000311186 | 997 (aa) | - | P16615 |
| ENST00000549840 | ATP2A2-208 | 503 | - | - | - (aa) | - | - |
| ENST00000552636 | ATP2A2-211 | 529 | - | ENSP00000447406 | 44 (aa) | - | F8W1Z7 |
| ENST00000377685 | ATP2A2-203 | 5702 | - | ENSP00000366913 | 50 (aa) | - | J3QSY6 |
| ENST00000548169 | ATP2A2-207 | 2951 | - | ENSP00000449454 | 933 (aa) | - | H7C5W9 |
| ENST00000550248 | ATP2A2-209 | 544 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04020 | Calcium signaling pathway | KEGG |
| hsa04022 | cGMP-PKG signaling pathway | KEGG |
| hsa04024 | cAMP signaling pathway | KEGG |
| hsa04260 | Cardiac muscle contraction | KEGG |
| hsa04261 | Adrenergic signaling in cardiomyocytes | KEGG |
| hsa04919 | Thyroid hormone signaling pathway | KEGG |
| hsa04972 | Pancreatic secretion | KEGG |
| hsa05010 | Alzheimer disease | KEGG |
| hsa05410 | Hypertrophic cardiomyopathy (HCM) | KEGG |
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | KEGG |
| hsa05414 | Dilated cardiomyopathy (DCM) | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000174437 | rs4766428 | 12 | 110285440 | T | Schizophrenia | 25056061 | [1.045-1.091] | 1.068 | EFO_0000692 |
| ENSG00000174437 | rs3026445 | 12 | 110285398 | C | QT interval | 24952745 | [0.44-0.8] unit increase | 0.62 | EFO_0004682 |
| ENSG00000174437 | rs3026485 | 12 | 110344222 | ? | Glucose homeostasis traits | 25524916 | [5.32-25.16] unit increase | 15.24 | EFO_0006896|EFO_0006831 |
| ENSG00000174437 | rs4766428 | 12 | 110285440 | T | Schizophrenia | 26198764 | [NR] | 1.07 | EFO_0000692 |
| ENSG00000174437 | rs4766428 | 12 | 110285440 | T | Schizophrenia | 28991256 | [1.04-1.09] | 1.065 | EFO_0000692 |
| ENSG00000174437 | rs4766428 | 12 | 110285440 | ? | Schizophrenia | 29483656 | [1.06-1.10] | 1.0787487 | EFO_0000692 |
| ENSG00000174437 | rs4766428 | 12 | 110285440 | T | Schizophrenia | 30285260 | [1.05-1.09] | 1.0719 | EFO_0000692 |
| ENSG00000174437 | rs4766428 | 12 | 110285440 | T | Schizophrenia | 30285260 | [1.05-1.09] | 1.07 | EFO_0000692 |
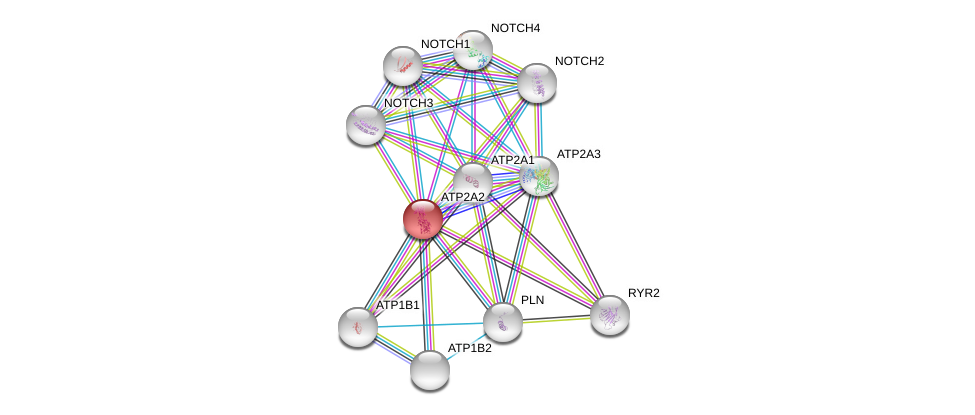
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000174437 | ATP2A2 | 100 | 98.898 | ENSMUSG00000029467 | Atp2a2 | 100 | 98.898 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002026 | regulation of the force of heart contraction | - | IEA | Process |
| GO:0003009 | skeletal muscle contraction | - | IEA | Process |
| GO:0005388 | calcium-transporting ATPase activity | 21873635. | IBA | Function |
| GO:0005388 | calcium-transporting ATPase activity | 16402920. | IDA | Function |
| GO:0005509 | calcium ion binding | 16402920. | IDA | Function |
| GO:0005515 | protein binding | 17526652.20528919.22084111.24764305.25996873.28298427. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005783 | endoplasmic reticulum | 16081076. | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | 11402072. | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | - | TAS | Component |
| GO:0005887 | integral component of plasma membrane | 2844796. | TAS | Component |
| GO:0006874 | cellular calcium ion homeostasis | 21873635. | IBA | Process |
| GO:0006984 | ER-nucleus signaling pathway | - | IEA | Process |
| GO:0006996 | organelle organization | - | IEA | Process |
| GO:0007155 | cell adhesion | 10080178. | TAS | Process |
| GO:0008544 | epidermis development | 10080178. | TAS | Process |
| GO:0008553 | proton-exporting ATPase activity, phosphorylative mechanism | 21873635. | IBA | Function |
| GO:0010460 | positive regulation of heart rate | 22679139. | TAS | Process |
| GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 21873635. | IBA | Process |
| GO:0012506 | vesicle membrane | - | IEA | Component |
| GO:0014883 | transition between fast and slow fiber | - | IEA | Process |
| GO:0014898 | cardiac muscle hypertrophy in response to stress | - | IEA | Process |
| GO:0016020 | membrane | 22375059. | IDA | Component |
| GO:0016529 | sarcoplasmic reticulum | 12804600. | IDA | Component |
| GO:0019899 | enzyme binding | 21903937. | IPI | Function |
| GO:0031095 | platelet dense tubular network membrane | - | TAS | Component |
| GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | - | IEA | Component |
| GO:0031775 | lutropin-choriogonadotropic hormone receptor binding | - | IEA | Function |
| GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 16402920. | IDA | Process |
| GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 16402920. | IDA | Process |
| GO:0032496 | response to lipopolysaccharide | - | IEA | Process |
| GO:0032991 | protein-containing complex | - | IEA | Component |
| GO:0033017 | sarcoplasmic reticulum membrane | 21873635. | IBA | Component |
| GO:0033017 | sarcoplasmic reticulum membrane | 19095005. | TAS | Component |
| GO:0033292 | T-tubule organization | - | IEA | Process |
| GO:0034220 | ion transmembrane transport | - | TAS | Process |
| GO:0034599 | cellular response to oxidative stress | - | IEA | Process |
| GO:0034605 | cellular response to heat | - | IEA | Process |
| GO:0034976 | response to endoplasmic reticulum stress | - | ISS | Process |
| GO:0044548 | S100 protein binding | 12804600. | IPI | Function |
| GO:0045822 | negative regulation of heart contraction | - | IEA | Process |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0055119 | relaxation of cardiac muscle | - | IEA | Process |
| GO:0061831 | apical ectoplasmic specialization | - | IEA | Component |
| GO:0070296 | sarcoplasmic reticulum calcium ion transport | 19095005.22679139. | TAS | Process |
| GO:0070588 | calcium ion transmembrane transport | 21873635. | IBA | Process |
| GO:0070588 | calcium ion transmembrane transport | 16402920. | IDA | Process |
| GO:0086036 | regulation of cardiac muscle cell membrane potential | 16402920. | IC | Process |
| GO:0086036 | regulation of cardiac muscle cell membrane potential | - | ISS | Process |
| GO:0086036 | regulation of cardiac muscle cell membrane potential | 22679139. | TAS | Process |
| GO:0086039 | calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential | 21873635. | IBA | Function |
| GO:0086039 | calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential | 16402920. | IC | Function |
| GO:0086039 | calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential | - | ISS | Function |
| GO:0086039 | calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential | 22679139. | TAS | Function |
| GO:0097470 | ribbon synapse | - | IEA | Component |
| GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction | - | ISS | Process |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | - | IEA | Process |
| GO:0120025 | plasma membrane bounded cell projection | - | IEA | Component |
| GO:1902600 | proton transmembrane transport | - | IEA | Process |
| GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter | - | IEA | Process |
| GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 16402920. | IDA | Process |
| GO:1903779 | regulation of cardiac conduction | - | TAS | Process |
| GO:1990036 | calcium ion import into sarcoplasmic reticulum | 16402920. | IC | Process |
| GO:1990036 | calcium ion import into sarcoplasmic reticulum | - | ISS | Process |