
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| KIRP | |||
| KIRP | |||
| LGG | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| SARC | ||
| MESO | ||
| SKCM | ||
| ESCA | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| KICH | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000518522 | SH3PXD2B-202 | 471 | - | ENSP00000428076 | 81 (aa) | - | H0YAU1 |
| ENST00000519643 | SH3PXD2B-203 | 1394 | - | ENSP00000430890 | 430 (aa) | - | G3V144 |
| ENST00000636523 | SH3PXD2B-205 | 1609 | - | ENSP00000490082 | 444 (aa) | - | A0A1B0GUF2 |
| ENST00000311601 | SH3PXD2B-201 | 7777 | XM_017009351 | ENSP00000309714 | 911 (aa) | XP_016864840 | A1X283 |
| ENST00000523651 | SH3PXD2B-204 | 390 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000174705 | Coronary Artery Disease | 7.6310000E-005 | - |
| ENSG00000174705 | Fibrinogen | 5.5960000E-005 | - |
| ENSG00000174705 | Child Development Disorders, Pervasive | 2.3552300E-005 | - |
| ENSG00000174705 | Child Development Disorders, Pervasive | 3.6261200E-005 | - |
| ENSG00000174705 | C-Reactive Protein | 7.3650000E-005 | - |
| ENSG00000174705 | C-Reactive Protein | 5.9890000E-005 | - |
| ENSG00000174705 | C-Reactive Protein | 5.1390000E-005 | - |
| ENSG00000174705 | C-Reactive Protein | 4.1060000E-005 | - |
| ENSG00000174705 | Body Mass Index | 9E-8 | 25133637 |
| ENSG00000174705 | Gastrointestinal Microbiome | 5E-8 | 27694959 |
| ENSG00000174705 | Body Mass Index | 5E-7 | 25133637 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000174705 | rs6866204 | 5 | 172426154 | ? | Balding type 1 | 30595370 | EFO_0007825 | ||
| ENSG00000174705 | rs6859752 | 5 | 172424930 | T | Male-pattern baldness | 30573740 | [0.026-0.039] unit decrease | 0.0328466 | EFO_0007825 |
| ENSG00000174705 | rs6866204 | 5 | 172426154 | ? | Waist-hip ratio | 30595370 | EFO_0004343 |
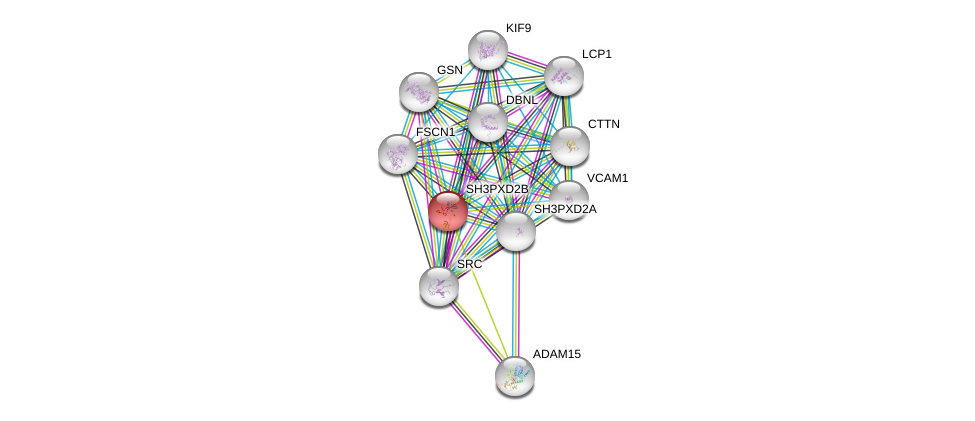
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001501 | skeletal system development | 20137777. | IMP | Process |
| GO:0001654 | eye development | 20137777. | IMP | Process |
| GO:0002051 | osteoblast fate commitment | - | IEA | Process |
| GO:0002102 | podosome | - | ISS | Component |
| GO:0005515 | protein binding | 19755710.20609497. | IPI | Function |
| GO:0005737 | cytoplasm | - | ISS | Component |
| GO:0006801 | superoxide metabolic process | 21873635. | IBA | Process |
| GO:0006801 | superoxide metabolic process | 19755710. | IDA | Process |
| GO:0007507 | heart development | 20137777. | IMP | Process |
| GO:0010314 | phosphatidylinositol-5-phosphate binding | 21873635. | IBA | Function |
| GO:0010314 | phosphatidylinositol-5-phosphate binding | - | ISS | Function |
| GO:0010628 | positive regulation of gene expression | - | IEA | Process |
| GO:0016176 | superoxide-generating NADPH oxidase activator activity | 21873635. | IBA | Function |
| GO:0022617 | extracellular matrix disassembly | 19755710. | IMP | Process |
| GO:0030054 | cell junction | - | IEA | Component |
| GO:0032266 | phosphatidylinositol-3-phosphate binding | 21873635. | IBA | Function |
| GO:0032266 | phosphatidylinositol-3-phosphate binding | - | ISS | Function |
| GO:0035091 | phosphatidylinositol binding | 21873635. | IBA | Function |
| GO:0040018 | positive regulation of multicellular organism growth | - | IEA | Process |
| GO:0042169 | SH2 domain binding | - | ISS | Function |
| GO:0042995 | cell projection | - | IEA | Component |
| GO:0043085 | positive regulation of catalytic activity | - | IEA | Process |
| GO:0045600 | positive regulation of fat cell differentiation | - | IEA | Process |
| GO:0048705 | skeletal system morphogenesis | - | IEA | Process |
| GO:0051496 | positive regulation of stress fiber assembly | - | IEA | Process |
| GO:0060348 | bone development | 20137777. | IMP | Process |
| GO:0060378 | regulation of brood size | - | IEA | Process |
| GO:0060612 | adipose tissue development | - | ISS | Process |
| GO:0070273 | phosphatidylinositol-4-phosphate binding | 21873635. | IBA | Function |
| GO:0071800 | podosome assembly | - | ISS | Process |
| GO:0072657 | protein localization to membrane | 19755710. | IDA | Process |
| GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding | 21873635. | IBA | Function |
| GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding | - | ISS | Function |
| GO:1904179 | positive regulation of adipose tissue development | - | IEA | Process |
| GO:1904888 | cranial skeletal system development | - | IEA | Process |