
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| KIRP | |||
| KIRP | |||
| LGG | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| KIRP | ||
| COAD | ||
| BLCA | ||
| UCEC | ||
| LIHC | ||
| CHOL | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000308496 | MMR_HSR1 | PF01926.23 | 7.8e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000310389 | ARL10-201 | 10845 | XM_011534531 | ENSP00000308496 | 244 (aa) | XP_011532833 | Q8N8L6 |
| ENST00000507151 | ARL10-203 | 365 | - | - | - (aa) | - | - |
| ENST00000503175 | ARL10-202 | 380 | - | ENSP00000424831 | 79 (aa) | - | H0Y9R6 |
| ENST00000514533 | ARL10-204 | 258 | - | ENSP00000421449 | 48 (aa) | - | H0Y8L6 |
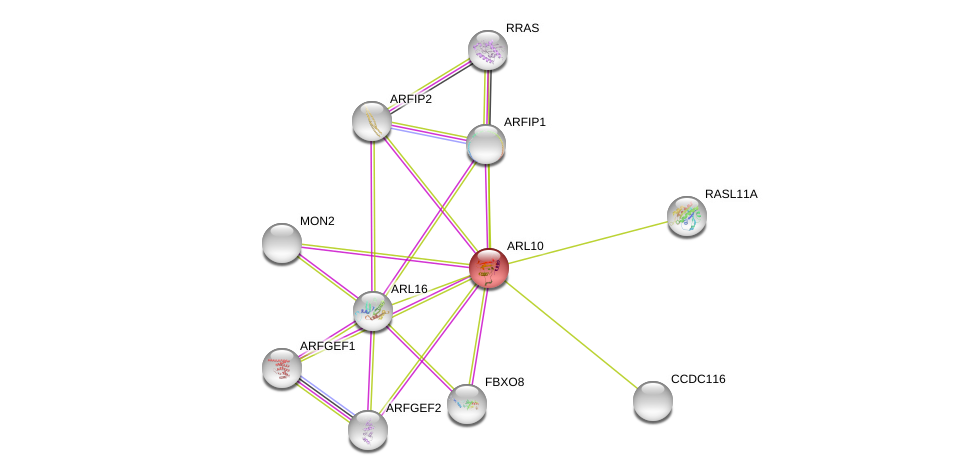
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000175414 | ARL10 | 99 | 92.562 | ENSAMEG00000006984 | ARL10 | 99 | 92.562 | Ailuropoda_melanoleuca |
| ENSG00000175414 | ARL10 | 92 | 65.909 | ENSACAG00000006350 | ARL10 | 91 | 45.259 | Anolis_carolinensis |
| ENSG00000175414 | ARL10 | 100 | 96.311 | ENSANAG00000029366 | ARL10 | 100 | 96.311 | Aotus_nancymaae |
| ENSG00000175414 | ARL10 | 94 | 93.333 | ENSBTAG00000025345 | ARL10 | 92 | 87.317 | Bos_taurus |
| ENSG00000175414 | ARL10 | 100 | 96.311 | ENSCJAG00000012332 | ARL10 | 100 | 96.311 | Callithrix_jacchus |
| ENSG00000175414 | ARL10 | 92 | 95.455 | ENSCAFG00000016697 | ARL10 | 87 | 94.048 | Canis_familiaris |
| ENSG00000175414 | ARL10 | 100 | 94.262 | ENSCAFG00020011500 | ARL10 | 100 | 94.262 | Canis_lupus_dingo |
| ENSG00000175414 | ARL10 | 69 | 90.909 | ENSTSYG00000036186 | ARL10 | 84 | 80.952 | Carlito_syrichta |
| ENSG00000175414 | ARL10 | 100 | 93.033 | ENSCPOG00000001642 | ARL10 | 100 | 93.033 | Cavia_porcellus |
| ENSG00000175414 | ARL10 | 100 | 95.902 | ENSCCAG00000032554 | ARL10 | 100 | 95.902 | Cebus_capucinus |
| ENSG00000175414 | ARL10 | 100 | 97.951 | ENSCATG00000012151 | ARL10 | 100 | 97.951 | Cercocebus_atys |
| ENSG00000175414 | ARL10 | 100 | 90.984 | ENSCLAG00000015048 | ARL10 | 100 | 90.984 | Chinchilla_lanigera |
| ENSG00000175414 | ARL10 | 100 | 98.770 | ENSCSAG00000009428 | ARL10 | 100 | 98.770 | Chlorocebus_sabaeus |
| ENSG00000175414 | ARL10 | 92 | 97.727 | ENSCANG00000036364 | ARL10 | 100 | 98.160 | Colobus_angolensis_palliatus |
| ENSG00000175414 | ARL10 | 100 | 88.525 | ENSCGRG00001022470 | - | 100 | 86.475 | Cricetulus_griseus_chok1gshd |
| ENSG00000175414 | ARL10 | 92 | 79.545 | ENSCGRG00000011571 | Arl10 | 89 | 88.095 | Cricetulus_griseus_crigri |
| ENSG00000175414 | ARL10 | 100 | 92.213 | ENSDNOG00000043008 | - | 100 | 92.213 | Dasypus_novemcinctus |
| ENSG00000175414 | ARL10 | 100 | 93.852 | ENSEASG00005011981 | ARL10 | 100 | 93.852 | Equus_asinus_asinus |
| ENSG00000175414 | ARL10 | 90 | 60.465 | ENSELUG00000011113 | - | 90 | 42.081 | Esox_lucius |
| ENSG00000175414 | ARL10 | 100 | 93.852 | ENSFCAG00000042295 | ARL10 | 100 | 93.852 | Felis_catus |
| ENSG00000175414 | ARL10 | 92 | 88.636 | ENSFDAG00000016410 | ARL10 | 84 | 91.071 | Fukomys_damarensis |
| ENSG00000175414 | ARL10 | 83 | 65.000 | ENSFHEG00000016443 | ARL10 | 88 | 39.367 | Fundulus_heteroclitus |
| ENSG00000175414 | ARL10 | 90 | 62.791 | ENSGMOG00000004201 | ARL10 | 98 | 50.370 | Gadus_morhua |
| ENSG00000175414 | ARL10 | 92 | 68.182 | ENSGALG00000003360 | ARL10 | 99 | 47.368 | Gallus_gallus |
| ENSG00000175414 | ARL10 | 94 | 71.111 | ENSGAGG00000013245 | ARL10 | 92 | 54.971 | Gopherus_agassizii |
| ENSG00000175414 | ARL10 | 100 | 99.590 | ENSGGOG00000005219 | - | 100 | 99.590 | Gorilla_gorilla |
| ENSG00000175414 | ARL10 | 97 | 88.186 | ENSHGLG00000005509 | ARL10 | 100 | 88.115 | Heterocephalus_glaber_female |
| ENSG00000175414 | ARL10 | 97 | 88.186 | ENSHGLG00100008434 | ARL10 | 100 | 88.115 | Heterocephalus_glaber_male |
| ENSG00000175414 | ARL10 | 100 | 91.020 | ENSSTOG00000004854 | ARL10 | 100 | 91.429 | Ictidomys_tridecemlineatus |
| ENSG00000175414 | ARL10 | 98 | 87.234 | ENSJJAG00000015006 | Arl10 | 100 | 79.508 | Jaculus_jaculus |
| ENSG00000175414 | ARL10 | 92 | 61.364 | ENSLACG00000014817 | ARL10 | 96 | 43.277 | Latimeria_chalumnae |
| ENSG00000175414 | ARL10 | 100 | 88.115 | ENSLAFG00000017880 | ARL10 | 100 | 88.115 | Loxodonta_africana |
| ENSG00000175414 | ARL10 | 100 | 98.361 | ENSMFAG00000033979 | ARL10 | 100 | 98.361 | Macaca_fascicularis |
| ENSG00000175414 | ARL10 | 100 | 98.361 | ENSMMUG00000019715 | ARL10 | 100 | 98.361 | Macaca_mulatta |
| ENSG00000175414 | ARL10 | 100 | 97.541 | ENSMNEG00000037218 | ARL10 | 100 | 97.541 | Macaca_nemestrina |
| ENSG00000175414 | ARL10 | 92 | 97.727 | ENSMLEG00000029969 | ARL10 | 91 | 96.739 | Mandrillus_leucophaeus |
| ENSG00000175414 | ARL10 | 100 | 86.475 | ENSMAUG00000016615 | Arl10 | 100 | 86.475 | Mesocricetus_auratus |
| ENSG00000175414 | ARL10 | 100 | 87.295 | ENSMOCG00000017185 | Arl10 | 100 | 86.475 | Microtus_ochrogaster |
| ENSG00000175414 | ARL10 | 94 | 84.444 | MGP_CAROLIEiJ_G0018537 | Arl10 | 92 | 86.096 | Mus_caroli |
| ENSG00000175414 | ARL10 | 94 | 84.444 | ENSMUSG00000025870 | Arl10 | 92 | 87.166 | Mus_musculus |
| ENSG00000175414 | ARL10 | 94 | 84.444 | MGP_PahariEiJ_G0019602 | Arl10 | 92 | 85.561 | Mus_pahari |
| ENSG00000175414 | ARL10 | 94 | 84.444 | MGP_SPRETEiJ_G0019417 | Arl10 | 92 | 87.166 | Mus_spretus |
| ENSG00000175414 | ARL10 | 100 | 93.852 | ENSMPUG00000012576 | ARL10 | 100 | 93.852 | Mustela_putorius_furo |
| ENSG00000175414 | ARL10 | 94 | 91.111 | ENSNGAG00000005101 | Arl10 | 81 | 91.979 | Nannospalax_galili |
| ENSG00000175414 | ARL10 | 98 | 93.617 | ENSNLEG00000012489 | - | 93 | 98.315 | Nomascus_leucogenys |
| ENSG00000175414 | ARL10 | 100 | 87.295 | ENSODEG00000009842 | ARL10 | 100 | 87.295 | Octodon_degus |
| ENSG00000175414 | ARL10 | 92 | 90.909 | ENSOCUG00000023334 | - | 87 | 91.071 | Oryctolagus_cuniculus |
| ENSG00000175414 | ARL10 | 96 | 63.043 | ENSORLG00000018307 | - | 89 | 44.118 | Oryzias_latipes |
| ENSG00000175414 | ARL10 | 90 | 65.116 | ENSOMEG00000023727 | ARL10 | 86 | 39.574 | Oryzias_melastigma |
| ENSG00000175414 | ARL10 | 100 | 90.574 | ENSOGAG00000000826 | ARL10 | 100 | 90.574 | Otolemur_garnettii |
| ENSG00000175414 | ARL10 | 92 | 95.455 | ENSOARG00000002098 | ARL10 | 100 | 79.508 | Ovis_aries |
| ENSG00000175414 | ARL10 | 92 | 100.000 | ENSPPAG00000039208 | - | 98 | 96.842 | Pan_paniscus |
| ENSG00000175414 | ARL10 | 100 | 93.852 | ENSPPRG00000017068 | ARL10 | 100 | 93.852 | Panthera_pardus |
| ENSG00000175414 | ARL10 | 92 | 95.455 | ENSPTIG00000011683 | ARL10 | 69 | 94.048 | Panthera_tigris_altaica |
| ENSG00000175414 | ARL10 | 100 | 99.590 | ENSPTRG00000046949 | ARL10 | 100 | 99.590 | Pan_troglodytes |
| ENSG00000175414 | ARL10 | 100 | 97.541 | ENSPANG00000011859 | ARL10 | 100 | 97.541 | Papio_anubis |
| ENSG00000175414 | ARL10 | 92 | 70.455 | ENSPSIG00000013641 | ARL10 | 97 | 55.090 | Pelodiscus_sinensis |
| ENSG00000175414 | ARL10 | 92 | 93.182 | ENSPCAG00000004681 | ARL10 | 100 | 81.557 | Procavia_capensis |
| ENSG00000175414 | ARL10 | 100 | 92.623 | ENSPCOG00000017730 | ARL10 | 100 | 92.623 | Propithecus_coquereli |
| ENSG00000175414 | ARL10 | 100 | 91.803 | ENSPVAG00000007499 | ARL10 | 100 | 91.803 | Pteropus_vampyrus |
| ENSG00000175414 | ARL10 | 92 | 88.636 | ENSRNOG00000045948 | Arl10 | 87 | 88.690 | Rattus_norvegicus |
| ENSG00000175414 | ARL10 | 100 | 98.770 | ENSRBIG00000030159 | ARL10 | 100 | 98.770 | Rhinopithecus_bieti |
| ENSG00000175414 | ARL10 | 100 | 98.770 | ENSRROG00000033178 | ARL10 | 100 | 98.770 | Rhinopithecus_roxellana |
| ENSG00000175414 | ARL10 | 71 | 84.483 | ENSSBOG00000031819 | ARL10 | 88 | 84.524 | Saimiri_boliviensis_boliviensis |
| ENSG00000175414 | ARL10 | 92 | 86.364 | ENSSHAG00000000707 | ARL10 | 99 | 62.753 | Sarcophilus_harrisii |
| ENSG00000175414 | ARL10 | 100 | 93.033 | ENSSSCG00000029985 | ARL10 | 100 | 93.033 | Sus_scrofa |
| ENSG00000175414 | ARL10 | 100 | 89.754 | ENSTTRG00000005396 | ARL10 | 100 | 89.754 | Tursiops_truncatus |
| ENSG00000175414 | ARL10 | 100 | 93.443 | ENSUAMG00000005136 | ARL10 | 100 | 93.443 | Ursus_americanus |
| ENSG00000175414 | ARL10 | 94 | 93.333 | ENSUMAG00000024507 | ARL10 | 90 | 74.684 | Ursus_maritimus |
| ENSG00000175414 | ARL10 | 100 | 94.262 | ENSVVUG00000001760 | ARL10 | 100 | 94.262 | Vulpes_vulpes |
| ENSG00000175414 | ARL10 | 85 | 65.854 | ENSXETG00000022337 | arl10 | 80 | 42.466 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005525 | GTP binding | - | IEA | Function |