
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| KIRP | |||
| KIRP | |||
| LGG | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| ACC | ||
| BRCA | ||
| GBM | ||
| LGG | ||
| CHOL |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000323714 | IF4E | PF01652.18 | 7.7e-53 | 1 | 1 |
| ENSP00000427633 | IF4E | PF01652.18 | 7.7e-53 | 1 | 1 |
| ENSP00000497422 | IF4E | PF01652.18 | 7.7e-53 | 1 | 1 |
| ENSP00000422420 | IF4E | PF01652.18 | 9.9e-33 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000505497 | EIF4E1B-204 | 514 | - | ENSP00000422420 | 171 (aa) | - | H0Y8X3 |
| ENST00000515458 | EIF4E1B-208 | 2836 | - | - | - (aa) | - | - |
| ENST00000318682 | EIF4E1B-201 | 1974 | XM_011534499 | ENSP00000323714 | 242 (aa) | XP_011532801 | A6NMX2 |
| ENST00000510660 | EIF4E1B-206 | 581 | - | ENSP00000421009 | 90 (aa) | - | D6RHE2 |
| ENST00000510473 | EIF4E1B-205 | 533 | - | - | - (aa) | - | - |
| ENST00000504597 | EIF4E1B-203 | 1676 | - | ENSP00000427633 | 242 (aa) | - | A6NMX2 |
| ENST00000647833 | EIF4E1B-209 | 2137 | - | ENSP00000497422 | 242 (aa) | - | UPI0001572CC7 |
| ENST00000503895 | EIF4E1B-202 | 564 | - | - | - (aa) | - | - |
| ENST00000512734 | EIF4E1B-207 | 561 | - | - | - (aa) | - | - |
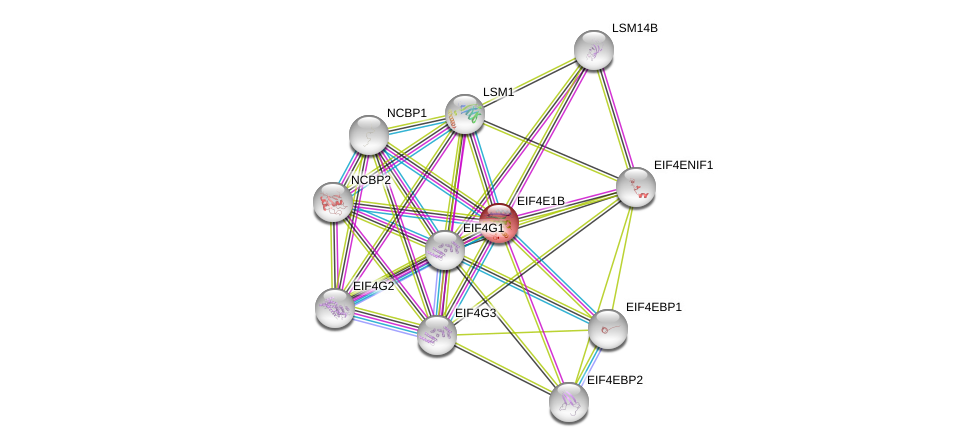
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000175766 | EIF4E1B | 75 | 74.033 | ENSG00000151247 | EIF4E | 83 | 74.033 |
| ENSG00000175766 | EIF4E1B | 93 | 31.602 | ENSG00000135930 | EIF4E2 | 84 | 34.574 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000175766 | EIF4E1B | 98 | 71.888 | ENSAMEG00000007196 | EIF4E1B | 85 | 77.103 | Ailuropoda_melanoleuca |
| ENSG00000175766 | EIF4E1B | 75 | 81.215 | ENSACAG00000007055 | EIF4E1B | 97 | 81.215 | Anolis_carolinensis |
| ENSG00000175766 | EIF4E1B | 100 | 89.256 | ENSANAG00000024720 | EIF4E1B | 100 | 88.843 | Aotus_nancymaae |
| ENSG00000175766 | EIF4E1B | 81 | 55.838 | ENSAMXG00000043293 | eif4e1b | 91 | 55.838 | Astyanax_mexicanus |
| ENSG00000175766 | EIF4E1B | 78 | 85.714 | ENSBTAG00000003103 | EIF4E1B | 82 | 85.714 | Bos_taurus |
| ENSG00000175766 | EIF4E1B | 100 | 88.843 | ENSCJAG00000007291 | EIF4E1B | 100 | 88.430 | Callithrix_jacchus |
| ENSG00000175766 | EIF4E1B | 97 | 82.979 | ENSCAFG00000016643 | EIF4E1B | 99 | 83.404 | Canis_familiaris |
| ENSG00000175766 | EIF4E1B | 75 | 91.713 | ENSCAFG00020011562 | EIF4E1B | 83 | 91.713 | Canis_lupus_dingo |
| ENSG00000175766 | EIF4E1B | 72 | 90.286 | ENSCHIG00000006321 | EIF4E1B | 90 | 90.286 | Capra_hircus |
| ENSG00000175766 | EIF4E1B | 100 | 70.248 | ENSTSYG00000025856 | EIF4E1B | 100 | 69.835 | Carlito_syrichta |
| ENSG00000175766 | EIF4E1B | 83 | 77.451 | ENSCAPG00000011182 | EIF4E1B | 91 | 77.451 | Cavia_aperea |
| ENSG00000175766 | EIF4E1B | 88 | 85.514 | ENSCPOG00000004429 | EIF4E1B | 97 | 80.753 | Cavia_porcellus |
| ENSG00000175766 | EIF4E1B | 100 | 90.083 | ENSCCAG00000008488 | EIF4E1B | 100 | 89.256 | Cebus_capucinus |
| ENSG00000175766 | EIF4E1B | 100 | 93.033 | ENSCATG00000036544 | EIF4E1B | 100 | 93.033 | Cercocebus_atys |
| ENSG00000175766 | EIF4E1B | 98 | 84.100 | ENSCLAG00000013384 | EIF4E1B | 92 | 90.625 | Chinchilla_lanigera |
| ENSG00000175766 | EIF4E1B | 100 | 90.283 | ENSCSAG00000009203 | EIF4E1B | 100 | 90.688 | Chlorocebus_sabaeus |
| ENSG00000175766 | EIF4E1B | 55 | 81.061 | ENSCPBG00000022627 | EIF4E1B | 100 | 81.061 | Chrysemys_picta_bellii |
| ENSG00000175766 | EIF4E1B | 75 | 62.431 | ENSCING00000004199 | - | 80 | 62.431 | Ciona_intestinalis |
| ENSG00000175766 | EIF4E1B | 100 | 93.033 | ENSCANG00000031424 | EIF4E1B | 100 | 94.672 | Colobus_angolensis_palliatus |
| ENSG00000175766 | EIF4E1B | 84 | 80.392 | ENSCGRG00001003887 | Eif4e1b | 96 | 88.811 | Cricetulus_griseus_chok1gshd |
| ENSG00000175766 | EIF4E1B | 75 | 72.376 | ENSDARG00000014565 | eif4e1b | 99 | 72.376 | Danio_rerio |
| ENSG00000175766 | EIF4E1B | 97 | 80.913 | ENSDNOG00000032189 | EIF4E1B | 77 | 88.235 | Dasypus_novemcinctus |
| ENSG00000175766 | EIF4E1B | 87 | 80.095 | ENSDORG00000016043 | Eif4e1b | 87 | 87.629 | Dipodomys_ordii |
| ENSG00000175766 | EIF4E1B | 72 | 61.714 | ENSEBUG00000014777 | eif4e1b | 85 | 61.714 | Eptatretus_burgeri |
| ENSG00000175766 | EIF4E1B | 99 | 83.951 | ENSEASG00005012081 | EIF4E1B | 99 | 83.951 | Equus_asinus_asinus |
| ENSG00000175766 | EIF4E1B | 98 | 83.750 | ENSECAG00000022357 | EIF4E1B | 87 | 84.583 | Equus_caballus |
| ENSG00000175766 | EIF4E1B | 75 | 77.348 | ENSEEUG00000005920 | EIF4E1B | 94 | 77.348 | Erinaceus_europaeus |
| ENSG00000175766 | EIF4E1B | 74 | 74.157 | ENSELUG00000021695 | eif4e1b | 76 | 74.157 | Esox_lucius |
| ENSG00000175766 | EIF4E1B | 99 | 81.048 | ENSFCAG00000003074 | EIF4E1B | 99 | 81.707 | Felis_catus |
| ENSG00000175766 | EIF4E1B | 75 | 81.215 | ENSFALG00000008607 | EIF4E1B | 82 | 81.215 | Ficedula_albicollis |
| ENSG00000175766 | EIF4E1B | 88 | 87.383 | ENSFDAG00000008704 | EIF4E1B | 98 | 81.743 | Fukomys_damarensis |
| ENSG00000175766 | EIF4E1B | 99 | 63.306 | ENSGALG00000026037 | EIF4E1B | 71 | 79.330 | Gallus_gallus |
| ENSG00000175766 | EIF4E1B | 59 | 82.517 | ENSGAGG00000012675 | EIF4E1B | 96 | 82.517 | Gopherus_agassizii |
| ENSG00000175766 | EIF4E1B | 100 | 98.889 | ENSGGOG00000015854 | EIF4E1B | 100 | 98.760 | Gorilla_gorilla |
| ENSG00000175766 | EIF4E1B | 88 | 89.252 | ENSHGLG00000016879 | - | 92 | 85.841 | Heterocephalus_glaber_female |
| ENSG00000175766 | EIF4E1B | 88 | 89.412 | ENSHGLG00100012839 | - | 97 | 89.412 | Heterocephalus_glaber_male |
| ENSG00000175766 | EIF4E1B | 75 | 74.586 | ENSIPUG00000020654 | eif4e1b | 92 | 70.707 | Ictalurus_punctatus |
| ENSG00000175766 | EIF4E1B | 98 | 79.253 | ENSSTOG00000002002 | EIF4E1B | 100 | 79.253 | Ictidomys_tridecemlineatus |
| ENSG00000175766 | EIF4E1B | 88 | 82.160 | ENSJJAG00000017541 | Eif4e1b | 87 | 82.160 | Jaculus_jaculus |
| ENSG00000175766 | EIF4E1B | 75 | 70.166 | ENSLOCG00000011832 | eif4e1b | 82 | 70.166 | Lepisosteus_oculatus |
| ENSG00000175766 | EIF4E1B | 73 | 78.212 | ENSLAFG00000027919 | EIF4E1B | 99 | 78.212 | Loxodonta_africana |
| ENSG00000175766 | EIF4E1B | 100 | 92.276 | ENSMFAG00000031726 | EIF4E1B | 99 | 96.682 | Macaca_fascicularis |
| ENSG00000175766 | EIF4E1B | 100 | 92.276 | ENSMMUG00000000142 | EIF4E1B | 100 | 92.683 | Macaca_mulatta |
| ENSG00000175766 | EIF4E1B | 100 | 92.276 | ENSMNEG00000031510 | EIF4E1B | 100 | 92.683 | Macaca_nemestrina |
| ENSG00000175766 | EIF4E1B | 100 | 92.276 | ENSMLEG00000037281 | EIF4E1B | 100 | 92.683 | Mandrillus_leucophaeus |
| ENSG00000175766 | EIF4E1B | 75 | 81.215 | ENSMGAG00000004602 | EIF4E1B | 83 | 81.215 | Meleagris_gallopavo |
| ENSG00000175766 | EIF4E1B | 100 | 69.478 | ENSMICG00000047427 | EIF4E1B | 100 | 69.880 | Microcebus_murinus |
| ENSG00000175766 | EIF4E1B | 50 | 83.471 | ENSMODG00000016941 | - | 95 | 83.471 | Monodelphis_domestica |
| ENSG00000175766 | EIF4E1B | 90 | 75.115 | ENSMODG00000005097 | - | 98 | 76.056 | Monodelphis_domestica |
| ENSG00000175766 | EIF4E1B | 97 | 70.638 | MGP_CAROLIEiJ_G0018546 | Eif4e1b | 100 | 70.085 | Mus_caroli |
| ENSG00000175766 | EIF4E1B | 91 | 72.727 | ENSMUSG00000074895 | Eif4e1b | 98 | 71.171 | Mus_musculus |
| ENSG00000175766 | EIF4E1B | 96 | 71.552 | MGP_PahariEiJ_G0019611 | Eif4e1b | 100 | 72.816 | Mus_pahari |
| ENSG00000175766 | EIF4E1B | 91 | 72.273 | MGP_SPRETEiJ_G0019426 | Eif4e1b | 98 | 71.171 | Mus_spretus |
| ENSG00000175766 | EIF4E1B | 98 | 81.405 | ENSMPUG00000012507 | EIF4E1B | 92 | 80.992 | Mustela_putorius_furo |
| ENSG00000175766 | EIF4E1B | 99 | 80.328 | ENSMLUG00000001437 | EIF4E1B | 85 | 87.143 | Myotis_lucifugus |
| ENSG00000175766 | EIF4E1B | 100 | 97.778 | ENSNLEG00000012532 | EIF4E1B | 100 | 97.107 | Nomascus_leucogenys |
| ENSG00000175766 | EIF4E1B | 73 | 71.186 | ENSMEUG00000002671 | EIF4E1B | 96 | 71.186 | Notamacropus_eugenii |
| ENSG00000175766 | EIF4E1B | 57 | 92.806 | ENSOPRG00000008875 | EIF4E1B | 93 | 92.806 | Ochotona_princeps |
| ENSG00000175766 | EIF4E1B | 97 | 79.079 | ENSODEG00000009390 | EIF4E1B | 99 | 79.079 | Octodon_degus |
| ENSG00000175766 | EIF4E1B | 76 | 91.848 | ENSOCUG00000007864 | EIF4E1B | 87 | 89.175 | Oryctolagus_cuniculus |
| ENSG00000175766 | EIF4E1B | 87 | 86.256 | ENSOGAG00000005844 | EIF4E1B | 100 | 86.256 | Otolemur_garnettii |
| ENSG00000175766 | EIF4E1B | 79 | 87.500 | ENSOARG00000002979 | EIF4E1B | 83 | 87.500 | Ovis_aries |
| ENSG00000175766 | EIF4E1B | 98 | 98.312 | ENSPPAG00000036530 | EIF4E1B | 95 | 98.312 | Pan_paniscus |
| ENSG00000175766 | EIF4E1B | 100 | 82.591 | ENSPPRG00000011119 | EIF4E1B | 100 | 81.781 | Panthera_pardus |
| ENSG00000175766 | EIF4E1B | 98 | 82.645 | ENSPTIG00000012304 | EIF4E1B | 99 | 82.231 | Panthera_tigris_altaica |
| ENSG00000175766 | EIF4E1B | 100 | 97.934 | ENSPTRG00000028481 | EIF4E1B | 100 | 97.934 | Pan_troglodytes |
| ENSG00000175766 | EIF4E1B | 100 | 91.870 | ENSPANG00000014775 | EIF4E1B | 100 | 92.276 | Papio_anubis |
| ENSG00000175766 | EIF4E1B | 75 | 72.928 | ENSPKIG00000025064 | eif4e1b | 85 | 72.928 | Paramormyrops_kingsleyae |
| ENSG00000175766 | EIF4E1B | 76 | 82.609 | ENSPCIG00000018442 | EIF4E1B | 72 | 82.609 | Phascolarctos_cinereus |
| ENSG00000175766 | EIF4E1B | 75 | 96.133 | ENSPPYG00000016077 | - | 83 | 96.133 | Pongo_abelii |
| ENSG00000175766 | EIF4E1B | 51 | 85.366 | ENSPCAG00000003618 | - | 100 | 85.366 | Procavia_capensis |
| ENSG00000175766 | EIF4E1B | 98 | 78.243 | ENSPCOG00000019904 | EIF4E1B | 99 | 78.903 | Propithecus_coquereli |
| ENSG00000175766 | EIF4E1B | 73 | 92.614 | ENSPVAG00000017039 | EIF4E1B | 96 | 81.731 | Pteropus_vampyrus |
| ENSG00000175766 | EIF4E1B | 75 | 73.481 | ENSPNAG00000000195 | eif4e1b | 84 | 73.481 | Pygocentrus_nattereri |
| ENSG00000175766 | EIF4E1B | 100 | 92.245 | ENSRBIG00000039242 | EIF4E1B | 99 | 96.682 | Rhinopithecus_bieti |
| ENSG00000175766 | EIF4E1B | 100 | 92.245 | ENSRROG00000042084 | EIF4E1B | 100 | 93.469 | Rhinopithecus_roxellana |
| ENSG00000175766 | EIF4E1B | 100 | 88.430 | ENSSBOG00000030710 | EIF4E1B | 100 | 88.017 | Saimiri_boliviensis_boliviensis |
| ENSG00000175766 | EIF4E1B | 76 | 82.703 | ENSSHAG00000007864 | EIF4E1B | 93 | 82.703 | Sarcophilus_harrisii |
| ENSG00000175766 | EIF4E1B | 81 | 69.744 | ENSSFOG00015005488 | eif4e1b | 89 | 69.744 | Scleropages_formosus |
| ENSG00000175766 | EIF4E1B | 54 | 79.348 | ENSSARG00000010425 | - | 83 | 79.592 | Sorex_araneus |
| ENSG00000175766 | EIF4E1B | 88 | 71.362 | ENSSPUG00000006304 | EIF4E1B | 99 | 79.545 | Sphenodon_punctatus |
| ENSG00000175766 | EIF4E1B | 98 | 78.601 | ENSSSCG00000014052 | EIF4E1B | 99 | 78.189 | Sus_scrofa |
| ENSG00000175766 | EIF4E1B | 69 | 80.723 | ENSTGUG00000000504 | EIF4E1B | 99 | 80.723 | Taeniopygia_guttata |
| ENSG00000175766 | EIF4E1B | 70 | 78.824 | ENSTBEG00000001631 | EIF4E1B | 94 | 78.824 | Tupaia_belangeri |
| ENSG00000175766 | EIF4E1B | 74 | 87.778 | ENSTTRG00000014026 | EIF4E1B | 85 | 87.778 | Tursiops_truncatus |
| ENSG00000175766 | EIF4E1B | 93 | 84.444 | ENSUAMG00000022136 | EIF4E1B | 98 | 88.462 | Ursus_americanus |
| ENSG00000175766 | EIF4E1B | 73 | 93.182 | ENSVVUG00000001475 | EIF4E1B | 73 | 93.182 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000340 | RNA 7-methylguanosine cap binding | 21873635. | IBA | Function |
| GO:0003743 | translation initiation factor activity | 21873635. | IBA | Function |
| GO:0005845 | mRNA cap binding complex | - | ISS | Component |
| GO:0006413 | translational initiation | - | IEA | Process |
| GO:0006417 | regulation of translation | - | IEA | Process |
| GO:0016281 | eukaryotic translation initiation factor 4F complex | 21873635. | IBA | Component |