
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| ESCA | |||
| ESCA | |||
| GBM | |||
| HNSC | |||
| LUAD | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| PCPG | |||
| PCPG | |||
| TGCT | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| UCEC | ||
| LIHC |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000533796 | TALDO1-209 | 503 | - | - | - (aa) | - | - |
| ENST00000532685 | TALDO1-208 | 568 | - | - | - (aa) | - | - |
| ENST00000530666 | TALDO1-206 | 587 | - | - | - (aa) | - | - |
| ENST00000528097 | TALDO1-203 | 1231 | - | ENSP00000437098 | 318 (aa) | - | F2Z393 |
| ENST00000530440 | TALDO1-205 | 710 | - | ENSP00000433501 | 93 (aa) | - | E9PKI8 |
| ENST00000319006 | TALDO1-201 | 1346 | - | ENSP00000321259 | 337 (aa) | - | P37837 |
| ENST00000530119 | TALDO1-204 | 1191 | - | - | - (aa) | - | - |
| ENST00000528070 | TALDO1-202 | 856 | - | ENSP00000435042 | 50 (aa) | - | E9PM01 |
| ENST00000532202 | TALDO1-207 | 270 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|
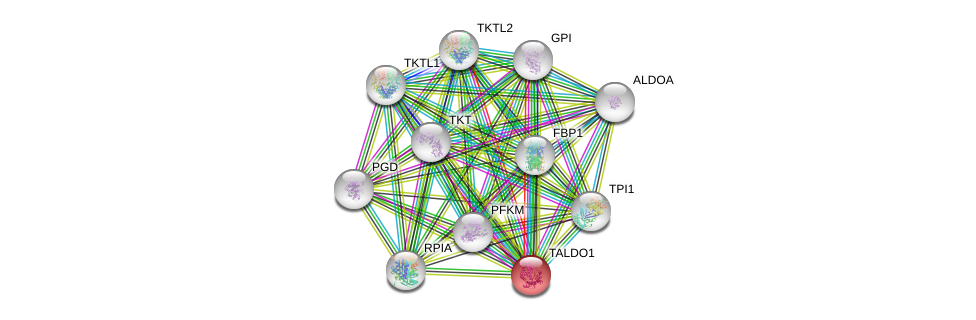
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000177156 | TALDO1 | 98 | 58.982 | YLR354C | 99 | 58.982 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004801 | sedoheptulose-7-phosphate:D-glyceraldehyde-3-phosphate glyceronetransferase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 21044950.25854864. | IPI | Function |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005737 | cytoplasm | 16130169. | TAS | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005975 | carbohydrate metabolic process | 9524206. | TAS | Process |
| GO:0005999 | xylulose biosynthetic process | - | TAS | Process |
| GO:0006002 | fructose 6-phosphate metabolic process | - | IEA | Process |
| GO:0006098 | pentose-phosphate shunt | - | TAS | Process |
| GO:0009052 | pentose-phosphate shunt, non-oxidative branch | 21873635. | IBA | Process |
| GO:0035722 | interleukin-12-mediated signaling pathway | - | TAS | Process |
| GO:0048029 | monosaccharide binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 20458337.23533145. | HDA | Component |