
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| BRCA | |||
| CHOL | |||
| HNSC | |||
| LIHC | |||
| LUAD | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| PRAD | |||
| SKCM | |||
| UCEC | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SKCM | ||
| COAD | ||
| PAAD | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| THCA | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000324560 | RIO1 | PF01163.22 | 0.00028 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000544718 | ULK1-207 | 1678 | - | - | - (aa) | - | - |
| ENST00000542313 | ULK1-206 | 307 | - | - | - (aa) | - | - |
| ENST00000321867 | ULK1-201 | 5310 | XM_011538798 | ENSP00000324560 | 1050 (aa) | XP_011537100 | O75385 |
| ENST00000537421 | ULK1-202 | 959 | - | - | - (aa) | - | - |
| ENST00000540568 | ULK1-203 | 2145 | - | - | - (aa) | - | - |
| ENST00000540647 | ULK1-204 | 1211 | - | - | - (aa) | - | - |
| ENST00000541761 | ULK1-205 | 1262 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000177169 | rs78661442 | 12 | 131920499 | ? | General cognitive ability | 29844566 | z-score increase | 4.433 | EFO_0004337 |
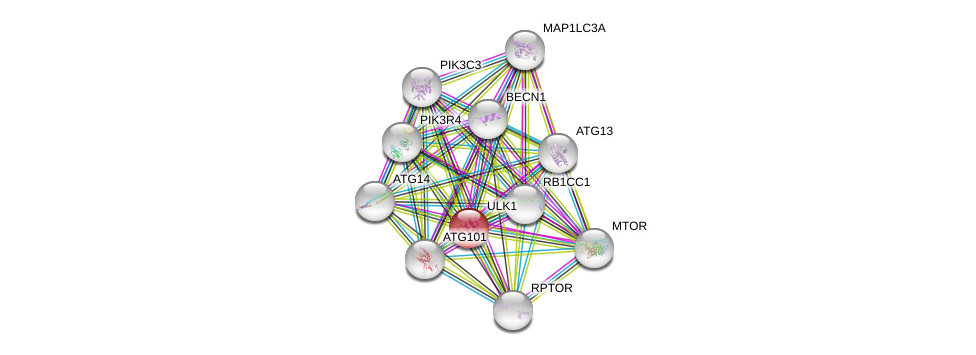
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000177169 | ULK1 | 97 | 83.137 | ENSAMEG00000013726 | ULK1 | 100 | 83.627 | Ailuropoda_melanoleuca |
| ENSG00000177169 | ULK1 | 97 | 74.206 | ENSAPLG00000014729 | ULK1 | 99 | 74.591 | Anas_platyrhynchos |
| ENSG00000177169 | ULK1 | 100 | 97.048 | ENSCJAG00000003693 | ULK1 | 100 | 97.048 | Callithrix_jacchus |
| ENSG00000177169 | ULK1 | 100 | 89.962 | ENSCHIG00000018901 | ULK1 | 100 | 90.436 | Capra_hircus |
| ENSG00000177169 | ULK1 | 96 | 90.659 | ENSTSYG00000032371 | ULK1 | 96 | 90.560 | Carlito_syrichta |
| ENSG00000177169 | ULK1 | 100 | 88.297 | ENSCPOG00000001024 | ULK1 | 100 | 88.297 | Cavia_porcellus |
| ENSG00000177169 | ULK1 | 100 | 93.175 | ENSCATG00000024995 | ULK1 | 100 | 93.340 | Cercocebus_atys |
| ENSG00000177169 | ULK1 | 100 | 97.714 | ENSCSAG00000001882 | ULK1 | 98 | 97.714 | Chlorocebus_sabaeus |
| ENSG00000177169 | ULK1 | 100 | 89.564 | ENSCGRG00001020729 | Ulk1 | 100 | 89.753 | Cricetulus_griseus_chok1gshd |
| ENSG00000177169 | ULK1 | 99 | 89.539 | ENSCGRG00000004904 | Ulk1 | 100 | 89.731 | Cricetulus_griseus_crigri |
| ENSG00000177169 | ULK1 | 100 | 90.814 | ENSEASG00005016522 | ULK1 | 100 | 91.004 | Equus_asinus_asinus |
| ENSG00000177169 | ULK1 | 100 | 90.814 | ENSECAG00000016158 | ULK1 | 100 | 91.440 | Equus_caballus |
| ENSG00000177169 | ULK1 | 100 | 89.489 | ENSFCAG00000010779 | ULK1 | 100 | 90.057 | Felis_catus |
| ENSG00000177169 | ULK1 | 99 | 71.895 | ENSFALG00000008276 | ULK1 | 98 | 72.217 | Ficedula_albicollis |
| ENSG00000177169 | ULK1 | 99 | 74.858 | ENSGALG00000002403 | ULK1 | 99 | 75.047 | Gallus_gallus |
| ENSG00000177169 | ULK1 | 98 | 73.007 | ENSGAGG00000002662 | ULK1 | 98 | 73.487 | Gopherus_agassizii |
| ENSG00000177169 | ULK1 | 100 | 87.216 | ENSHGLG00000016386 | ULK1 | 100 | 87.879 | Heterocephalus_glaber_female |
| ENSG00000177169 | ULK1 | 99 | 83.789 | ENSLAFG00000003503 | ULK1 | 99 | 84.166 | Loxodonta_africana |
| ENSG00000177169 | ULK1 | 100 | 97.714 | ENSMNEG00000034338 | ULK1 | 100 | 97.714 | Macaca_nemestrina |
| ENSG00000177169 | ULK1 | 100 | 92.381 | ENSMICG00000011757 | ULK1 | 100 | 92.476 | Microcebus_murinus |
| ENSG00000177169 | ULK1 | 100 | 88.994 | ENSMOCG00000014550 | Ulk1 | 100 | 89.374 | Microtus_ochrogaster |
| ENSG00000177169 | ULK1 | 99 | 75.283 | ENSMODG00000015566 | ULK1 | 99 | 75.189 | Monodelphis_domestica |
| ENSG00000177169 | ULK1 | 100 | 89.089 | MGP_CAROLIEiJ_G0027552 | Ulk1 | 100 | 89.279 | Mus_caroli |
| ENSG00000177169 | ULK1 | 100 | 89.279 | ENSMUSG00000029512 | Ulk1 | 100 | 89.469 | Mus_musculus |
| ENSG00000177169 | ULK1 | 100 | 89.005 | MGP_PahariEiJ_G0024134 | Ulk1 | 100 | 89.194 | Mus_pahari |
| ENSG00000177169 | ULK1 | 100 | 89.279 | MGP_SPRETEiJ_G0028550 | Ulk1 | 100 | 89.469 | Mus_spretus |
| ENSG00000177169 | ULK1 | 100 | 89.848 | ENSNGAG00000007610 | Ulk1 | 100 | 90.038 | Nannospalax_galili |
| ENSG00000177169 | ULK1 | 100 | 98.190 | ENSNLEG00000008460 | ULK1 | 100 | 98.190 | Nomascus_leucogenys |
| ENSG00000177169 | ULK1 | 84 | 86.885 | ENSOPRG00000002632 | - | 84 | 86.885 | Ochotona_princeps |
| ENSG00000177169 | ULK1 | 100 | 87.347 | ENSODEG00000008683 | ULK1 | 100 | 87.347 | Octodon_degus |
| ENSG00000177169 | ULK1 | 99 | 74.388 | ENSOANG00000013859 | ULK1 | 99 | 74.388 | Ornithorhynchus_anatinus |
| ENSG00000177169 | ULK1 | 99 | 89.364 | ENSOCUG00000015070 | ULK1 | 99 | 89.364 | Oryctolagus_cuniculus |
| ENSG00000177169 | ULK1 | 100 | 92.762 | ENSOGAG00000005056 | ULK1 | 100 | 92.762 | Otolemur_garnettii |
| ENSG00000177169 | ULK1 | 98 | 99.420 | ENSPPAG00000043077 | - | 100 | 99.420 | Pan_paniscus |
| ENSG00000177169 | ULK1 | 97 | 70.452 | ENSPSIG00000008801 | ULK1 | 100 | 70.837 | Pelodiscus_sinensis |
| ENSG00000177169 | ULK1 | 99 | 89.037 | ENSPEMG00000010074 | Ulk1 | 100 | 89.228 | Peromyscus_maniculatus_bairdii |
| ENSG00000177169 | ULK1 | 99 | 78.544 | ENSPCIG00000006060 | ULK1 | 99 | 78.450 | Phascolarctos_cinereus |
| ENSG00000177169 | ULK1 | 85 | 85.762 | ENSPCAG00000004814 | ULK1 | 100 | 86.652 | Procavia_capensis |
| ENSG00000177169 | ULK1 | 100 | 93.048 | ENSPCOG00000001314 | ULK1 | 100 | 93.143 | Propithecus_coquereli |
| ENSG00000177169 | ULK1 | 100 | 88.899 | ENSRNOG00000037505 | Ulk1 | 100 | 89.089 | Rattus_norvegicus |
| ENSG00000177169 | ULK1 | 100 | 97.333 | ENSRBIG00000042231 | ULK1 | 100 | 97.333 | Rhinopithecus_bieti |
| ENSG00000177169 | ULK1 | 99 | 77.996 | ENSSHAG00000013399 | ULK1 | 99 | 77.715 | Sarcophilus_harrisii |
| ENSG00000177169 | ULK1 | 100 | 90.246 | ENSSSCG00000009742 | ULK1 | 100 | 90.720 | Sus_scrofa |
| ENSG00000177169 | ULK1 | 93 | 68.844 | ENSXETG00000002028 | ulk1 | 99 | 69.447 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000045 | autophagosome assembly | 21873635. | IBA | Process |
| GO:0000407 | phagophore assembly site | 21873635. | IBA | Component |
| GO:0004674 | protein serine/threonine kinase activity | 21873635. | IBA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 18936157.25126726.27103069. | IDA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 9693035. | NAS | Function |
| GO:0004674 | protein serine/threonine kinase activity | - | TAS | Function |
| GO:0005515 | protein binding | 11146101.17389358.18936157.19211835.19597335.20562859.21072212.22354037.22613832.23392225.23524951.24290141.24603492.25126726.25127057.25438055.25686248.25891078.26347139.26593251.27334615.28195531.28561066.29950492. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005737 | cytoplasm | 9693035. | NAS | Component |
| GO:0005741 | mitochondrial outer membrane | - | TAS | Component |
| GO:0005776 | autophagosome | 17595159.22456507. | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | - | TAS | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | 11146101. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006468 | protein phosphorylation | 9693035. | NAS | Process |
| GO:0006914 | autophagy | 18936157. | IDA | Process |
| GO:0006914 | autophagy | 17595159. | IMP | Process |
| GO:0006914 | autophagy | - | ISS | Process |
| GO:0007165 | signal transduction | - | IEA | Process |
| GO:0008104 | protein localization | 16940348. | IMP | Process |
| GO:0010506 | regulation of autophagy | 21873635. | IBA | Process |
| GO:0016020 | membrane | 21873635. | IBA | Component |
| GO:0016236 | macroautophagy | 22354037. | IMP | Process |
| GO:0016236 | macroautophagy | - | TAS | Process |
| GO:0016241 | regulation of macroautophagy | - | TAS | Process |
| GO:0016301 | kinase activity | - | TAS | Function |
| GO:0017137 | Rab GTPase binding | 22613832. | IPI | Function |
| GO:0018105 | peptidyl-serine phosphorylation | 25126726.27103069. | IDA | Process |
| GO:0018107 | peptidyl-threonine phosphorylation | 27103069. | IDA | Process |
| GO:0030424 | axon | - | IEA | Component |
| GO:0031102 | neuron projection regeneration | - | IEA | Process |
| GO:0031175 | neuron projection development | 18007665. | IMP | Process |
| GO:0031333 | negative regulation of protein complex assembly | 25126726. | IDA | Process |
| GO:0031669 | cellular response to nutrient levels | - | ISS | Process |
| GO:0032045 | guanyl-nucleotide exchange factor complex | 27103069. | IDA | Component |
| GO:0034045 | phagophore assembly site membrane | 18936157. | IDA | Component |
| GO:0042594 | response to starvation | - | ISS | Process |
| GO:0042802 | identical protein binding | 20562859.23524951. | IPI | Function |
| GO:0044877 | protein-containing complex binding | 19211835. | IPI | Function |
| GO:0046777 | protein autophosphorylation | 21873635. | IBA | Process |
| GO:0046777 | protein autophosphorylation | 18936157. | IDA | Process |
| GO:0048675 | axon extension | 21873635. | IBA | Process |
| GO:0051020 | GTPase binding | 25891078. | IPI | Function |
| GO:0055037 | recycling endosome | - | TAS | Component |
| GO:0075044 | autophagy of host cells involved in interaction with symbiont | 28389568. | IGI | Process |
| GO:0097629 | extrinsic component of omegasome membrane | 22885598. | IDA | Component |
| GO:0097632 | extrinsic component of phagophore assembly site membrane | 22885598. | IDA | Component |
| GO:0097635 | extrinsic component of autophagosome membrane | 22885598. | IDA | Component |
| GO:1990316 | Atg1/ULK1 kinase complex | 19211835. | IPI | Component |
| GO:2000786 | positive regulation of autophagosome assembly | - | IEA | Process |