
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| BRCA | |||
| DLBC | |||
| THCA | |||
| UCEC | |||
| UCS | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| SARC | ||
| MESO | ||
| HNSC | ||
| LUSC | ||
| ESCA | ||
| COAD | ||
| BLCA | ||
| READ | ||
| GBM | ||
| THCA | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000357037 | CAVIN1-201 | 3828 | - | ENSP00000349541 | 390 (aa) | - | Q6NZI2 |
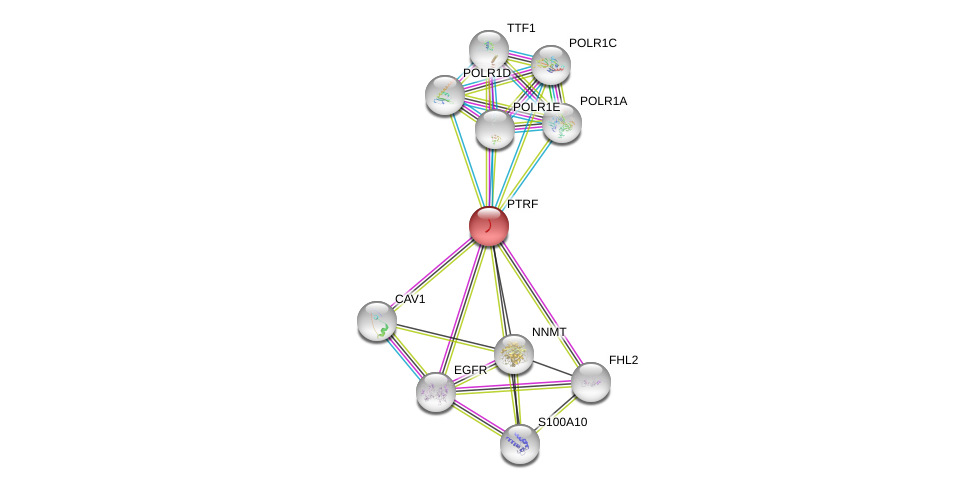
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000177469 | CAVIN1 | 81 | 41.945 | ENSMUSG00000045954 | Cavin2 | 73 | 38.375 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005515 | protein binding | 17026959.19525939.24013648.24567387. | IPI | Function |
| GO:0005634 | nucleus | 15242332. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 17026959. | IDA | Component |
| GO:0005739 | mitochondrion | 15242332. | IDA | Component |
| GO:0005783 | endoplasmic reticulum | - | IEA | Component |
| GO:0005829 | cytosol | - | IEA | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0005901 | caveola | 21873635. | IBA | Component |
| GO:0005901 | caveola | 17026959.19525939.24013648. | IDA | Component |
| GO:0006361 | transcription initiation from RNA polymerase I promoter | 9582279. | IDA | Process |
| GO:0006363 | termination of RNA polymerase I transcription | 9582279. | IDA | Process |
| GO:0006363 | termination of RNA polymerase I transcription | - | TAS | Process |
| GO:0009303 | rRNA transcription | - | ISS | Process |
| GO:0009306 | protein secretion | - | IEA | Process |
| GO:0032991 | protein-containing complex | 24013648. | IDA | Component |
| GO:0042134 | rRNA primary transcript binding | 9582279. | IDA | Function |
| GO:0042802 | identical protein binding | 24013648. | IPI | Function |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0045121 | membrane raft | 25204797. | IDA | Component |
| GO:2000147 | positive regulation of cell motility | - | ISS | Process |