
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| PAAD | ||
| BLCA | ||
| READ | ||
| LAML | ||
| LGG | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000480851 | cwf21 | PF08312.12 | 2.7e-14 | 1 | 1 |
| ENSP00000480851 | SRRM_C | PF15230.6 | 1.8e-23 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000612155 | SRRM3-205 | 2150 | - | - | - (aa) | - | - |
| ENST00000479294 | SRRM3-203 | 549 | - | - | - (aa) | - | - |
| ENST00000464752 | SRRM3-201 | 2797 | - | ENSP00000483698 | 109 (aa) | - | A0A087X0W5 |
| ENST00000611745 | SRRM3-204 | 3612 | - | ENSP00000480851 | 653 (aa) | - | A0A087WXA3 |
| ENST00000479284 | SRRM3-202 | 589 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000177679 | rs11770797 | 7 | 76226193 | ? | Blood protein levels | 28240269 | EFO_0008242 | ||
| ENSG00000177679 | rs4728638 | 7 | 76272712 | T | Educational attainment (MTAG) | 30038396 | [0.0083-0.0157] unit increase | 0.012 | EFO_0004784 |
| ENSG00000177679 | rs3960554 | 7 | 76216279 | G | Eotaxin levels | 27989323 | [0.047-0.113] SD units decrease | 0.0798 | EFO_0008122 |
| ENSG00000177679 | rs60717745 | 7 | 76216765 | C | Educational attainment (years of education) | 30038396 | [0.0081-0.0155] unit decrease | 0.0118 | EFO_0004784 |
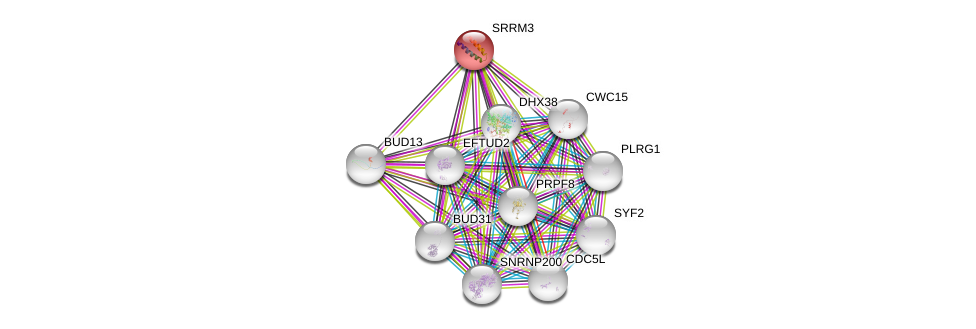
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000177679 | SRRM3 | 87 | 51.178 | ENSACAG00000000452 | SRRM3 | 51 | 64.605 | Anolis_carolinensis |
| ENSG00000177679 | SRRM3 | 73 | 98.750 | ENSANAG00000031983 | SRRM3 | 59 | 85.326 | Aotus_nancymaae |
| ENSG00000177679 | SRRM3 | 61 | 90.909 | ENSBTAG00000007879 | - | 98 | 84.639 | Bos_taurus |
| ENSG00000177679 | SRRM3 | 100 | 96.172 | ENSCJAG00000016307 | SRRM3 | 100 | 96.172 | Callithrix_jacchus |
| ENSG00000177679 | SRRM3 | 95 | 79.936 | ENSCAFG00000013537 | SRRM3 | 73 | 92.388 | Canis_familiaris |
| ENSG00000177679 | SRRM3 | 81 | 76.136 | ENSCAFG00020013073 | SRRM3 | 88 | 77.108 | Canis_lupus_dingo |
| ENSG00000177679 | SRRM3 | 100 | 96.784 | ENSCCAG00000033359 | SRRM3 | 100 | 96.784 | Cebus_capucinus |
| ENSG00000177679 | SRRM3 | 89 | 85.204 | ENSCLAG00000007682 | SRRM3 | 100 | 91.129 | Chinchilla_lanigera |
| ENSG00000177679 | SRRM3 | 73 | 98.750 | ENSCSAG00000018140 | SRRM3 | 97 | 96.546 | Chlorocebus_sabaeus |
| ENSG00000177679 | SRRM3 | 89 | 87.671 | ENSCGRG00001011385 | Srrm3 | 51 | 90.361 | Cricetulus_griseus_chok1gshd |
| ENSG00000177679 | SRRM3 | 100 | 73.893 | ENSDNOG00000046922 | SRRM3 | 56 | 69.780 | Dasypus_novemcinctus |
| ENSG00000177679 | SRRM3 | 72 | 93.750 | ENSEASG00005016840 | - | 88 | 93.093 | Equus_asinus_asinus |
| ENSG00000177679 | SRRM3 | 75 | 91.463 | ENSECAG00000022142 | - | 88 | 92.771 | Equus_caballus |
| ENSG00000177679 | SRRM3 | 63 | 94.203 | ENSFCAG00000044112 | SRRM3 | 55 | 89.227 | Felis_catus |
| ENSG00000177679 | SRRM3 | 95 | 53.282 | ENSFDAG00000016913 | SRRM3 | 99 | 97.500 | Fukomys_damarensis |
| ENSG00000177679 | SRRM3 | 70 | 91.262 | ENSGGOG00000036497 | - | 82 | 90.244 | Gorilla_gorilla |
| ENSG00000177679 | SRRM3 | 83 | 91.346 | ENSSTOG00000014181 | SRRM3 | 100 | 91.129 | Ictidomys_tridecemlineatus |
| ENSG00000177679 | SRRM3 | 99 | 81.860 | ENSJJAG00000023441 | Srrm3 | 60 | 78.237 | Jaculus_jaculus |
| ENSG00000177679 | SRRM3 | 58 | 57.513 | ENSMGAG00000003690 | SRRM3 | 75 | 64.014 | Meleagris_gallopavo |
| ENSG00000177679 | SRRM3 | 89 | 83.390 | ENSMAUG00000017613 | Srrm3 | 100 | 89.516 | Mesocricetus_auratus |
| ENSG00000177679 | SRRM3 | 88 | 92.857 | ENSMICG00000010668 | SRRM3 | 56 | 93.072 | Microcebus_murinus |
| ENSG00000177679 | SRRM3 | 96 | 88.198 | ENSMOCG00000002565 | Srrm3 | 51 | 89.458 | Microtus_ochrogaster |
| ENSG00000177679 | SRRM3 | 61 | 92.424 | MGP_CAROLIEiJ_G0027845 | - | 97 | 90.361 | Mus_caroli |
| ENSG00000177679 | SRRM3 | 96 | 88.836 | ENSMUSG00000039860 | Srrm3 | 100 | 90.323 | Mus_musculus |
| ENSG00000177679 | SRRM3 | 95 | 88.350 | MGP_PahariEiJ_G0024425 | Srrm3 | 100 | 89.516 | Mus_pahari |
| ENSG00000177679 | SRRM3 | 96 | 88.836 | MGP_SPRETEiJ_G0028844 | Srrm3 | 100 | 91.129 | Mus_spretus |
| ENSG00000177679 | SRRM3 | 100 | 72.129 | ENSNLEG00000008423 | - | 54 | 80.422 | Nomascus_leucogenys |
| ENSG00000177679 | SRRM3 | 89 | 89.384 | ENSODEG00000015988 | SRRM3 | 56 | 89.227 | Octodon_degus |
| ENSG00000177679 | SRRM3 | 61 | 87.879 | ENSOGAG00000030805 | SRRM3 | 55 | 91.667 | Otolemur_garnettii |
| ENSG00000177679 | SRRM3 | 100 | 98.928 | ENSPTRG00000019317 | SRRM3 | 100 | 98.928 | Pan_troglodytes |
| ENSG00000177679 | SRRM3 | 70 | 79.339 | ENSPCAG00000013700 | SRRM3 | 76 | 79.339 | Procavia_capensis |
| ENSG00000177679 | SRRM3 | 100 | 91.118 | ENSPCOG00000028464 | SRRM3 | 55 | 91.160 | Propithecus_coquereli |
| ENSG00000177679 | SRRM3 | 58 | 78.796 | ENSPVAG00000003400 | SRRM3 | 55 | 92.606 | Pteropus_vampyrus |
| ENSG00000177679 | SRRM3 | 96 | 88.995 | ENSRNOG00000001439 | Srrm3 | 51 | 89.759 | Rattus_norvegicus |
| ENSG00000177679 | SRRM3 | 79 | 96.951 | ENSRROG00000034980 | SRRM3 | 94 | 96.951 | Rhinopithecus_roxellana |
| ENSG00000177679 | SRRM3 | 85 | 45.161 | ENSSPUG00000006285 | SRRM3 | 65 | 78.182 | Sphenodon_punctatus |
| ENSG00000177679 | SRRM3 | 61 | 85.124 | ENSTTRG00000014836 | - | 67 | 85.124 | Tursiops_truncatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003729 | mRNA binding | 21873635. | IBA | Function |