
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| ESCA | |||
| READ | |||
| SARC | |||
| THCA | |||
| UCEC | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| PRAD | ||
| LUSC | ||
| ESCA | ||
| COAD | ||
| CESC | ||
| LAML | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000327031 | FLII-201 | 4338 | XM_005256555 | ENSP00000324573 | 1269 (aa) | XP_005256612 | Q13045 |
| ENST00000579294 | FLII-222 | 3936 | - | ENSP00000463534 | 1258 (aa) | - | Q13045 |
| ENST00000584444 | FLII-229 | 782 | - | - | - (aa) | - | - |
| ENST00000577398 | FLII-216 | 926 | - | ENSP00000468045 | 207 (aa) | - | K7EQZ7 |
| ENST00000577626 | FLII-219 | 557 | - | ENSP00000463902 | 117 (aa) | - | J3QQU5 |
| ENST00000628188 | FLII-230 | 156 | - | ENSP00000486754 | 51 (aa) | - | J3KS39 |
| ENST00000487369 | FLII-208 | 522 | - | - | - (aa) | - | - |
| ENST00000488932 | FLII-211 | 838 | - | ENSP00000463838 | 279 (aa) | - | J3QQQ2 |
| ENST00000473425 | FLII-205 | 585 | - | ENSP00000467212 | 99 (aa) | - | K7EP37 |
| ENST00000578101 | FLII-220 | 4148 | - | ENSP00000462293 | 51 (aa) | - | J3KS39 |
| ENST00000580453 | FLII-223 | 673 | - | - | - (aa) | - | - |
| ENST00000578558 | FLII-221 | 2377 | - | ENSP00000462314 | 700 (aa) | - | J3KS54 |
| ENST00000496727 | FLII-214 | 792 | - | - | - (aa) | - | - |
| ENST00000582626 | FLII-228 | 573 | - | - | - (aa) | - | - |
| ENST00000581858 | FLII-227 | 644 | - | ENSP00000465202 | 23 (aa) | - | K7EJJ7 |
| ENST00000493600 | FLII-213 | 656 | - | - | - (aa) | - | - |
| ENST00000488221 | FLII-210 | 859 | - | - | - (aa) | - | - |
| ENST00000474265 | FLII-206 | 596 | - | - | - (aa) | - | - |
| ENST00000581349 | FLII-225 | 582 | - | ENSP00000467199 | 133 (aa) | - | K7EP27 |
| ENST00000577402 | FLII-217 | 689 | - | - | - (aa) | - | - |
| ENST00000580966 | FLII-224 | 514 | - | - | - (aa) | - | - |
| ENST00000493401 | FLII-212 | 458 | - | - | - (aa) | - | - |
| ENST00000465046 | FLII-204 | 699 | - | - | - (aa) | - | - |
| ENST00000581401 | FLII-226 | 385 | - | ENSP00000462797 | 29 (aa) | - | J3KT47 |
| ENST00000461110 | FLII-203 | 1032 | - | - | - (aa) | - | - |
| ENST00000478416 | FLII-207 | 878 | - | - | - (aa) | - | - |
| ENST00000459958 | FLII-202 | 692 | - | - | - (aa) | - | - |
| ENST00000577485 | FLII-218 | 555 | - | ENSP00000463675 | 185 (aa) | - | J3QLR6 |
| ENST00000545457 | FLII-215 | 3975 | - | ENSP00000438536 | 1214 (aa) | - | Q13045 |
| ENST00000487693 | FLII-209 | 669 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000177731 | rs68165736 | 17 | 18253846 | ? | Hair color | 30595370 | EFO_0007822 |
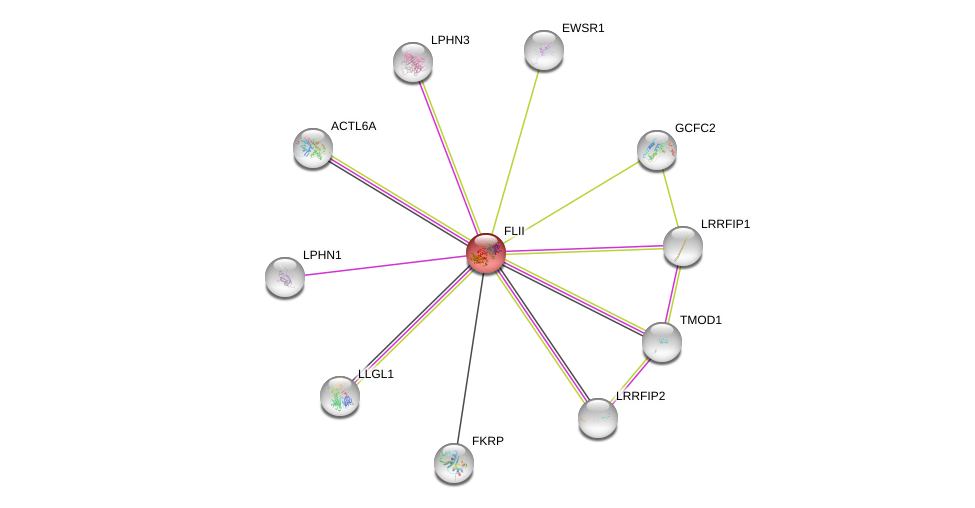
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003779 | actin binding | 9525888. | IDA | Function |
| GO:0005515 | protein binding | 9671805.10366446. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005815 | microtubule organizing center | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005903 | brush border | - | IEA | Component |
| GO:0005925 | focal adhesion | - | IEA | Component |
| GO:0007275 | multicellular organism development | - | IEA | Process |
| GO:0030036 | actin cytoskeleton organization | - | IEA | Process |
| GO:0051014 | actin filament severing | - | IEA | Process |
| GO:0051015 | actin filament binding | - | IEA | Function |