
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| CESC | |||
| GBM | |||
| KIRP | |||
| LGG | |||
| LUSC | |||
| SARC | |||
| SKCM | |||
| THCA | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| SARC | ||
| SKCM | ||
| PRAD | ||
| TGCT | ||
| COAD | ||
| PAAD | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000392564 | GRB2-205 | 3182 | - | ENSP00000376347 | 217 (aa) | - | P62993 |
| ENST00000392562 | GRB2-203 | 3729 | - | ENSP00000376345 | 217 (aa) | - | P62993 |
| ENST00000392563 | GRB2-204 | 2851 | - | ENSP00000376346 | 176 (aa) | - | P62993 |
| ENST00000577711 | GRB2-207 | 555 | - | - | - (aa) | - | - |
| ENST00000462266 | GRB2-206 | 601 | - | - | - (aa) | - | - |
| ENST00000581959 | GRB2-209 | 359 | - | ENSP00000463525 | 42 (aa) | - | J3QLF6 |
| ENST00000316804 | GRB2-202 | 3248 | - | ENSP00000339007 | 217 (aa) | - | P62993 |
| ENST00000316615 | GRB2-201 | 3181 | - | ENSP00000317360 | 176 (aa) | - | P62993 |
| ENST00000648046 | GRB2-212 | 1711 | - | ENSP00000496913 | 217 (aa) | - | B0LPF3 |
| ENST00000582582 | GRB2-210 | 584 | - | ENSP00000462785 | 110 (aa) | - | J3KT38 |
| ENST00000583912 | GRB2-211 | 561 | - | - | - (aa) | - | - |
| ENST00000578961 | GRB2-208 | 687 | - | ENSP00000464279 | 156 (aa) | - | J3QRL5 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | KEGG |
| hsa01522 | Endocrine resistance | KEGG |
| hsa04010 | MAPK signaling pathway | KEGG |
| hsa04012 | ErbB signaling pathway | KEGG |
| hsa04014 | Ras signaling pathway | KEGG |
| hsa04062 | Chemokine signaling pathway | KEGG |
| hsa04068 | FoxO signaling pathway | KEGG |
| hsa04072 | Phospholipase D signaling pathway | KEGG |
| hsa04150 | mTOR signaling pathway | KEGG |
| hsa04151 | PI3K-Akt signaling pathway | KEGG |
| hsa04380 | Osteoclast differentiation | KEGG |
| hsa04510 | Focal adhesion | KEGG |
| hsa04540 | Gap junction | KEGG |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | KEGG |
| hsa04630 | JAK-STAT signaling pathway | KEGG |
| hsa04650 | Natural killer cell mediated cytotoxicity | KEGG |
| hsa04660 | T cell receptor signaling pathway | KEGG |
| hsa04662 | B cell receptor signaling pathway | KEGG |
| hsa04664 | Fc epsilon RI signaling pathway | KEGG |
| hsa04714 | Thermogenesis | KEGG |
| hsa04722 | Neurotrophin signaling pathway | KEGG |
| hsa04910 | Insulin signaling pathway | KEGG |
| hsa04912 | GnRH signaling pathway | KEGG |
| hsa04915 | Estrogen signaling pathway | KEGG |
| hsa04917 | Prolactin signaling pathway | KEGG |
| hsa04926 | Relaxin signaling pathway | KEGG |
| hsa05034 | Alcoholism | KEGG |
| hsa05160 | Hepatitis C | KEGG |
| hsa05161 | Hepatitis B | KEGG |
| hsa05163 | Human cytomegalovirus infection | KEGG |
| hsa05165 | Human papillomavirus infection | KEGG |
| hsa05200 | Pathways in cancer | KEGG |
| hsa05203 | Viral carcinogenesis | KEGG |
| hsa05205 | Proteoglycans in cancer | KEGG |
| hsa05206 | MicroRNAs in cancer | KEGG |
| hsa05210 | Colorectal cancer | KEGG |
| hsa05211 | Renal cell carcinoma | KEGG |
| hsa05213 | Endometrial cancer | KEGG |
| hsa05214 | Glioma | KEGG |
| hsa05215 | Prostate cancer | KEGG |
| hsa05220 | Chronic myeloid leukemia | KEGG |
| hsa05221 | Acute myeloid leukemia | KEGG |
| hsa05223 | Non-small cell lung cancer | KEGG |
| hsa05224 | Breast cancer | KEGG |
| hsa05225 | Hepatocellular carcinoma | KEGG |
| hsa05226 | Gastric cancer | KEGG |
| hsa05231 | Choline metabolism in cancer | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000177885 | rs9894206 | 17 | 75364073 | ? | Systemic lupus erythematosus | 28714469 | [1.15-1.39] | 1.25 | EFO_0002690 |
| ENSG00000177885 | rs36023980 | 17 | 75345203 | C | Systemic lupus erythematosus | 29848360 | [1.13-1.24] | 1.18 | EFO_0002690 |
| ENSG00000177885 | rs2385263 | 17 | 75325036 | ? | Waist-hip ratio | 30595370 | EFO_0004343 |
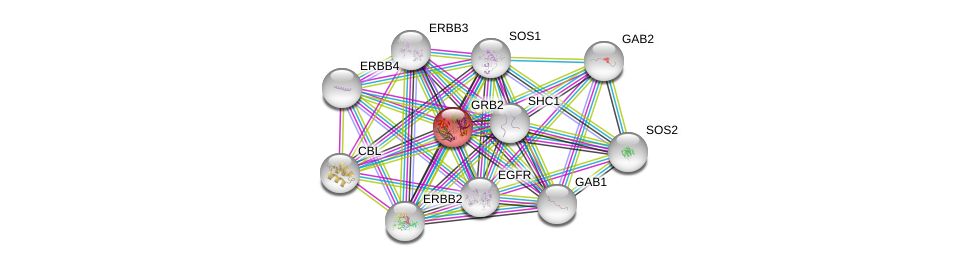
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000177885 | GRB2 | 100 | 100.000 | ENSMUSG00000059923 | Grb2 | 100 | 100.000 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000165 | MAPK cascade | - | TAS | Process |
| GO:0001784 | phosphotyrosine residue binding | 20624904. | IPI | Function |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005070 | SH3/SH2 adaptor activity | 8253073. | TAS | Function |
| GO:0005154 | epidermal growth factor receptor binding | 12577067. | IPI | Function |
| GO:0005168 | neurotrophin TRKA receptor binding | 15488758. | IPI | Function |
| GO:0005515 | protein binding | 7504175.7518560.7518772.7522233.7523381.7527391.7537265.7629138.7773779.8112292.8491186.8663178.8798570.8816475.8887653.8892607.8939605.8940013.8955135.8959326.8995379.9020117.9045636.9148935.9174058.9175774.9178760.9178913.9254595.9346925.9447984.9489702.9544989.9569023.9788432.10026169.10206341.10229072.10318918.10377264.10660596.10660620.10811803.10887132.10980613.11278465.11432792.11684012.11741929.11827484.11882361.11896612.11923305.12091307.12213123.12359715.12577067.14679214.14722116.14993658.15467741.15558030.15657067.15784897.15929943.16253990.16273093.16374509.16407827.16520382.16696976.16730941.16799092.16893902.16899217.16912232.16938345.17094785.17235283.17276458.17474147.18273061.18624398.18647389.18654987.18692475.19172738.19323566.19380743.19509291.19615732.19798056.19914172.20933011.20936779.21179510.21189249.21278786.21278788.21706016.21725282.21725358.21822214.21900206.21930792.21988832.22323290.22536782.22726438.23178716.23460737.23799367.24189400.24658140.24728074.25036637.25092874.25416956.25535246.25814554.25819436.27059931.27107012.28046066.28330616.29997244. | IPI | Function |
| GO:0005634 | nucleus | 21179510. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005730 | nucleolus | - | IDA | Component |
| GO:0005737 | cytoplasm | 21179510. | IDA | Component |
| GO:0005768 | endosome | 21179510. | IDA | Component |
| GO:0005794 | Golgi apparatus | - | IEA | Component |
| GO:0005829 | cytosol | 14722116. | TAS | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005911 | cell-cell junction | - | IEA | Component |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 1322798. | TAS | Process |
| GO:0007265 | Ras protein signal transduction | 8253073. | TAS | Process |
| GO:0007267 | cell-cell signaling | 8253073. | TAS | Process |
| GO:0007411 | axon guidance | - | TAS | Process |
| GO:0007568 | aging | - | IEA | Process |
| GO:0008180 | COP9 signalosome | 22561606. | IDA | Component |
| GO:0008286 | insulin receptor signaling pathway | 8388384. | IPI | Process |
| GO:0008286 | insulin receptor signaling pathway | - | TAS | Process |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | - | TAS | Process |
| GO:0012506 | vesicle membrane | - | IEA | Component |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0017124 | SH3 domain binding | 19509291. | IDA | Function |
| GO:0019221 | cytokine-mediated signaling pathway | - | TAS | Process |
| GO:0019901 | protein kinase binding | 20086093. | IPI | Function |
| GO:0019903 | protein phosphatase binding | - | IEA | Function |
| GO:0030838 | positive regulation of actin filament polymerization | - | IEA | Process |
| GO:0031295 | T cell costimulation | - | TAS | Process |
| GO:0031623 | receptor internalization | 14985334. | IMP | Process |
| GO:0035635 | entry of bacterium into host cell | - | TAS | Process |
| GO:0035723 | interleukin-15-mediated signaling pathway | - | TAS | Process |
| GO:0038095 | Fc-epsilon receptor signaling pathway | - | TAS | Process |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | - | TAS | Process |
| GO:0038128 | ERBB2 signaling pathway | - | TAS | Process |
| GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | - | TAS | Process |
| GO:0042770 | signal transduction in response to DNA damage | 20160708. | IMP | Process |
| GO:0042802 | identical protein binding | 12577067.22536782.22726438. | IPI | Function |
| GO:0043408 | regulation of MAPK cascade | - | IEA | Process |
| GO:0043560 | insulin receptor substrate binding | 8388384. | IPI | Function |
| GO:0046579 | positive regulation of Ras protein signal transduction | - | TAS | Process |
| GO:0046875 | ephrin receptor binding | 12925710. | IPI | Function |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | - | TAS | Process |
| GO:0048646 | anatomical structure formation involved in morphogenesis | - | IEA | Process |
| GO:0050900 | leukocyte migration | - | TAS | Process |
| GO:0051291 | protein heterooligomerization | - | IEA | Process |
| GO:0051897 | positive regulation of protein kinase B signaling | - | TAS | Process |
| GO:0060670 | branching involved in labyrinthine layer morphogenesis | - | IEA | Process |
| GO:0061024 | membrane organization | - | TAS | Process |
| GO:0070062 | extracellular exosome | 19056867.20458337.23533145. | HDA | Component |
| GO:0070436 | Grb2-EGFR complex | 20086093. | IDA | Component |
| GO:0071479 | cellular response to ionizing radiation | 20160708. | IMP | Process |
| GO:2000379 | positive regulation of reactive oxygen species metabolic process | 20160708. | IMP | Process |