
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CESC | |||
| COAD | |||
| GBM | |||
| HNSC | |||
| KICH | |||
| KIRP | |||
| LUAD | |||
| MESO | |||
| PAAD | |||
| READ | |||
| SARC | |||
| STAD | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| ACC | ||
| KIRP | ||
| COAD | ||
| PCPG | ||
| BLCA | ||
| CESC | ||
| LIHC | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000321029 | DNA_pol_A_exo1 | PF01612.20 | 1.8e-09 | 1 | 1 |
| ENSP00000415056 | DNA_pol_A_exo1 | PF01612.20 | 2.2e-09 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000558881 | EXD1-204 | 2293 | - | - | - (aa) | - | - |
| ENST00000558396 | EXD1-203 | 483 | - | ENSP00000452678 | 55 (aa) | - | H0YK66 |
| ENST00000314992 | EXD1-201 | 2942 | XM_011521299 | ENSP00000321029 | 514 (aa) | XP_011519601 | Q8NHP7 |
| ENST00000559743 | EXD1-205 | 601 | - | - | - (aa) | - | - |
| ENST00000458580 | EXD1-202 | 2177 | XM_006720415 | ENSP00000415056 | 572 (aa) | XP_006720478 | Q8NHP7 |
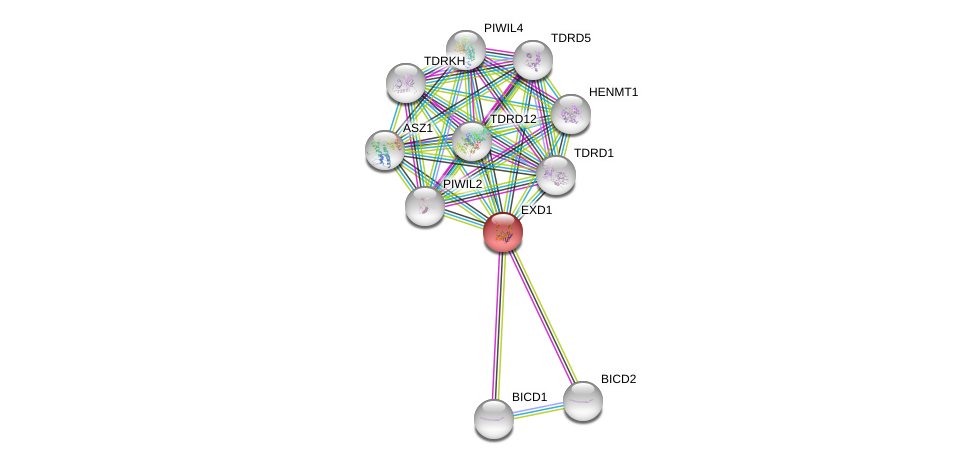
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000178997 | EXD1 | 54 | 46.290 | ENSAPOG00000004287 | - | 68 | 52.709 | Acanthochromis_polyacanthus |
| ENSG00000178997 | EXD1 | 99 | 80.035 | ENSAMEG00000016793 | EXD1 | 99 | 80.035 | Ailuropoda_melanoleuca |
| ENSG00000178997 | EXD1 | 73 | 47.500 | ENSAOCG00000009355 | - | 71 | 44.554 | Amphiprion_ocellaris |
| ENSG00000178997 | EXD1 | 73 | 47.500 | ENSAPEG00000002194 | - | 86 | 46.154 | Amphiprion_percula |
| ENSG00000178997 | EXD1 | 89 | 53.061 | ENSACAG00000003676 | EXD1 | 100 | 57.975 | Anolis_carolinensis |
| ENSG00000178997 | EXD1 | 100 | 90.227 | ENSANAG00000029058 | EXD1 | 100 | 90.227 | Aotus_nancymaae |
| ENSG00000178997 | EXD1 | 70 | 40.842 | ENSACLG00000001361 | - | 72 | 40.988 | Astatotilapia_calliptera |
| ENSG00000178997 | EXD1 | 75 | 48.780 | ENSAMXG00000026016 | - | 75 | 41.278 | Astyanax_mexicanus |
| ENSG00000178997 | EXD1 | 92 | 80.632 | ENSBTAG00000008363 | EXD1 | 100 | 80.249 | Bos_taurus |
| ENSG00000178997 | EXD1 | 99 | 84.790 | ENSCJAG00000020765 | EXD1 | 99 | 84.790 | Callithrix_jacchus |
| ENSG00000178997 | EXD1 | 99 | 79.826 | ENSCAFG00000009510 | EXD1 | 100 | 79.826 | Canis_familiaris |
| ENSG00000178997 | EXD1 | 82 | 78.873 | ENSCAFG00020005504 | EXD1 | 100 | 78.873 | Canis_lupus_dingo |
| ENSG00000178997 | EXD1 | 99 | 77.113 | ENSCHIG00000019606 | EXD1 | 99 | 77.113 | Capra_hircus |
| ENSG00000178997 | EXD1 | 98 | 81.481 | ENSCPOG00000010573 | EXD1 | 100 | 74.520 | Cavia_porcellus |
| ENSG00000178997 | EXD1 | 100 | 87.260 | ENSCCAG00000016905 | EXD1 | 100 | 87.260 | Cebus_capucinus |
| ENSG00000178997 | EXD1 | 100 | 94.942 | ENSCATG00000037400 | EXD1 | 100 | 94.942 | Cercocebus_atys |
| ENSG00000178997 | EXD1 | 98 | 75.049 | ENSCSAG00000007647 | EXD1 | 100 | 75.049 | Chlorocebus_sabaeus |
| ENSG00000178997 | EXD1 | 98 | 74.308 | ENSCHOG00000011441 | EXD1 | 99 | 74.308 | Choloepus_hoffmanni |
| ENSG00000178997 | EXD1 | 99 | 52.840 | ENSCPBG00000013511 | EXD1 | 93 | 53.012 | Chrysemys_picta_bellii |
| ENSG00000178997 | EXD1 | 100 | 91.783 | ENSCANG00000036848 | EXD1 | 100 | 91.783 | Colobus_angolensis_palliatus |
| ENSG00000178997 | EXD1 | 99 | 75.868 | ENSCGRG00001011355 | Exd1 | 97 | 75.868 | Cricetulus_griseus_chok1gshd |
| ENSG00000178997 | EXD1 | 99 | 75.868 | ENSCGRG00000011633 | Exd1 | 99 | 75.868 | Cricetulus_griseus_crigri |
| ENSG00000178997 | EXD1 | 67 | 41.818 | ENSCSEG00000000904 | - | 68 | 41.818 | Cynoglossus_semilaevis |
| ENSG00000178997 | EXD1 | 63 | 36.034 | ENSDARG00000098669 | exd1 | 85 | 40.684 | Danio_rerio |
| ENSG00000178997 | EXD1 | 100 | 80.836 | ENSDNOG00000011410 | EXD1 | 100 | 80.314 | Dasypus_novemcinctus |
| ENSG00000178997 | EXD1 | 84 | 86.957 | ENSDORG00000012079 | Exd1 | 100 | 74.382 | Dipodomys_ordii |
| ENSG00000178997 | EXD1 | 78 | 70.556 | ENSETEG00000014257 | - | 79 | 70.556 | Echinops_telfairi |
| ENSG00000178997 | EXD1 | 98 | 83.333 | ENSEASG00005016394 | EXD1 | 100 | 80.139 | Equus_asinus_asinus |
| ENSG00000178997 | EXD1 | 100 | 82.056 | ENSECAG00000021250 | EXD1 | 100 | 82.056 | Equus_caballus |
| ENSG00000178997 | EXD1 | 98 | 71.937 | ENSEEUG00000001232 | EXD1 | 100 | 71.937 | Erinaceus_europaeus |
| ENSG00000178997 | EXD1 | 70 | 42.680 | ENSELUG00000006324 | - | 67 | 42.822 | Esox_lucius |
| ENSG00000178997 | EXD1 | 99 | 82.548 | ENSFCAG00000028533 | EXD1 | 99 | 82.548 | Felis_catus |
| ENSG00000178997 | EXD1 | 56 | 45.897 | ENSGMOG00000015956 | - | 96 | 45.593 | Gadus_morhua |
| ENSG00000178997 | EXD1 | 68 | 53.827 | ENSGALG00000041449 | EXD1 | 96 | 53.571 | Gallus_gallus |
| ENSG00000178997 | EXD1 | 70 | 41.855 | ENSGAFG00000017161 | - | 72 | 41.353 | Gambusia_affinis |
| ENSG00000178997 | EXD1 | 99 | 52.405 | ENSGAGG00000019200 | EXD1 | 93 | 52.405 | Gopherus_agassizii |
| ENSG00000178997 | EXD1 | 100 | 98.833 | ENSGGOG00000004594 | EXD1 | 100 | 98.833 | Gorilla_gorilla |
| ENSG00000178997 | EXD1 | 50 | 48.649 | ENSHBUG00000017880 | - | 64 | 49.421 | Haplochromis_burtoni |
| ENSG00000178997 | EXD1 | 100 | 76.614 | ENSHGLG00000015369 | EXD1 | 100 | 76.614 | Heterocephalus_glaber_female |
| ENSG00000178997 | EXD1 | 99 | 75.524 | ENSHGLG00100007398 | EXD1 | 100 | 75.524 | Heterocephalus_glaber_male |
| ENSG00000178997 | EXD1 | 100 | 83.595 | ENSSTOG00000014980 | EXD1 | 100 | 83.595 | Ictidomys_tridecemlineatus |
| ENSG00000178997 | EXD1 | 75 | 40.835 | ENSLBEG00000000022 | - | 73 | 41.067 | Labrus_bergylta |
| ENSG00000178997 | EXD1 | 89 | 38.776 | ENSLACG00000006680 | EXD1 | 69 | 50.360 | Latimeria_chalumnae |
| ENSG00000178997 | EXD1 | 82 | 46.667 | ENSLOCG00000012278 | - | 96 | 45.000 | Lepisosteus_oculatus |
| ENSG00000178997 | EXD1 | 99 | 77.104 | ENSLAFG00000014739 | EXD1 | 100 | 78.236 | Loxodonta_africana |
| ENSG00000178997 | EXD1 | 100 | 95.455 | ENSMFAG00000034670 | EXD1 | 100 | 95.455 | Macaca_fascicularis |
| ENSG00000178997 | EXD1 | 100 | 95.105 | ENSMMUG00000013991 | EXD1 | 100 | 95.105 | Macaca_mulatta |
| ENSG00000178997 | EXD1 | 100 | 95.629 | ENSMNEG00000037333 | EXD1 | 100 | 95.629 | Macaca_nemestrina |
| ENSG00000178997 | EXD1 | 100 | 98.182 | ENSMLEG00000033153 | EXD1 | 100 | 97.279 | Mandrillus_leucophaeus |
| ENSG00000178997 | EXD1 | 70 | 41.089 | ENSMZEG00005009193 | - | 75 | 41.728 | Maylandia_zebra |
| ENSG00000178997 | EXD1 | 78 | 48.837 | ENSMGAG00000011179 | EXD1 | 99 | 53.133 | Meleagris_gallopavo |
| ENSG00000178997 | EXD1 | 99 | 75.524 | ENSMAUG00000002698 | Exd1 | 99 | 75.524 | Mesocricetus_auratus |
| ENSG00000178997 | EXD1 | 100 | 82.927 | ENSMICG00000014337 | EXD1 | 100 | 82.927 | Microcebus_murinus |
| ENSG00000178997 | EXD1 | 99 | 73.951 | ENSMOCG00000018151 | Exd1 | 99 | 73.951 | Microtus_ochrogaster |
| ENSG00000178997 | EXD1 | 98 | 57.143 | ENSMODG00000000164 | EXD1 | 100 | 65.085 | Monodelphis_domestica |
| ENSG00000178997 | EXD1 | 70 | 40.099 | ENSMALG00000015814 | - | 86 | 40.695 | Monopterus_albus |
| ENSG00000178997 | EXD1 | 98 | 79.630 | MGP_CAROLIEiJ_G0024145 | Exd1 | 99 | 73.997 | Mus_caroli |
| ENSG00000178997 | EXD1 | 99 | 73.252 | ENSMUSG00000048647 | Exd1 | 99 | 73.252 | Mus_musculus |
| ENSG00000178997 | EXD1 | 98 | 79.630 | MGP_PahariEiJ_G0025588 | Exd1 | 99 | 68.476 | Mus_pahari |
| ENSG00000178997 | EXD1 | 99 | 73.601 | MGP_SPRETEiJ_G0025061 | Exd1 | 99 | 73.601 | Mus_spretus |
| ENSG00000178997 | EXD1 | 99 | 81.533 | ENSMPUG00000008811 | EXD1 | 99 | 81.533 | Mustela_putorius_furo |
| ENSG00000178997 | EXD1 | 99 | 80.196 | ENSMLUG00000001466 | EXD1 | 100 | 80.377 | Myotis_lucifugus |
| ENSG00000178997 | EXD1 | 99 | 75.874 | ENSNGAG00000002319 | Exd1 | 99 | 75.874 | Nannospalax_galili |
| ENSG00000178997 | EXD1 | 52 | 49.813 | ENSNBRG00000003229 | - | 72 | 49.813 | Neolamprologus_brichardi |
| ENSG00000178997 | EXD1 | 100 | 96.109 | ENSNLEG00000003456 | EXD1 | 100 | 96.109 | Nomascus_leucogenys |
| ENSG00000178997 | EXD1 | 99 | 65.900 | ENSOPRG00000016433 | EXD1 | 100 | 65.900 | Ochotona_princeps |
| ENSG00000178997 | EXD1 | 68 | 41.730 | ENSONIG00000012043 | - | 98 | 41.985 | Oreochromis_niloticus |
| ENSG00000178997 | EXD1 | 98 | 57.407 | ENSOANG00000029781 | - | 91 | 51.601 | Ornithorhynchus_anatinus |
| ENSG00000178997 | EXD1 | 98 | 74.354 | ENSOCUG00000013817 | EXD1 | 99 | 75.049 | Oryctolagus_cuniculus |
| ENSG00000178997 | EXD1 | 67 | 42.708 | ENSORLG00000026344 | - | 76 | 40.988 | Oryzias_latipes |
| ENSG00000178997 | EXD1 | 67 | 43.490 | ENSORLG00020005465 | - | 73 | 43.490 | Oryzias_latipes_hni |
| ENSG00000178997 | EXD1 | 67 | 42.448 | ENSORLG00015004848 | - | 73 | 41.860 | Oryzias_latipes_hsok |
| ENSG00000178997 | EXD1 | 67 | 43.411 | ENSOMEG00000019812 | - | 77 | 41.565 | Oryzias_melastigma |
| ENSG00000178997 | EXD1 | 99 | 75.534 | ENSOGAG00000013095 | EXD1 | 100 | 75.240 | Otolemur_garnettii |
| ENSG00000178997 | EXD1 | 99 | 81.261 | ENSOARG00000020330 | EXD1 | 100 | 80.936 | Ovis_aries |
| ENSG00000178997 | EXD1 | 100 | 98.951 | ENSPPAG00000033760 | EXD1 | 100 | 98.951 | Pan_paniscus |
| ENSG00000178997 | EXD1 | 99 | 81.501 | ENSPPRG00000011128 | EXD1 | 99 | 81.501 | Panthera_pardus |
| ENSG00000178997 | EXD1 | 99 | 81.326 | ENSPTIG00000020026 | EXD1 | 99 | 81.326 | Panthera_tigris_altaica |
| ENSG00000178997 | EXD1 | 100 | 99.126 | ENSPTRG00000006941 | EXD1 | 100 | 99.126 | Pan_troglodytes |
| ENSG00000178997 | EXD1 | 100 | 95.280 | ENSPANG00000019000 | EXD1 | 100 | 95.280 | Papio_anubis |
| ENSG00000178997 | EXD1 | 68 | 41.837 | ENSPKIG00000000671 | - | 70 | 41.221 | Paramormyrops_kingsleyae |
| ENSG00000178997 | EXD1 | 98 | 51.399 | ENSPSIG00000014473 | EXD1 | 92 | 51.399 | Pelodiscus_sinensis |
| ENSG00000178997 | EXD1 | 70 | 37.531 | ENSPMGG00000015466 | - | 78 | 37.531 | Periophthalmus_magnuspinnatus |
| ENSG00000178997 | EXD1 | 89 | 77.551 | ENSPCIG00000027845 | EXD1 | 100 | 65.400 | Phascolarctos_cinereus |
| ENSG00000178997 | EXD1 | 69 | 41.191 | ENSPFOG00000013673 | - | 97 | 39.951 | Poecilia_formosa |
| ENSG00000178997 | EXD1 | 70 | 41.791 | ENSPLAG00000014262 | - | 72 | 41.294 | Poecilia_latipinna |
| ENSG00000178997 | EXD1 | 70 | 41.542 | ENSPMEG00000012071 | - | 72 | 41.045 | Poecilia_mexicana |
| ENSG00000178997 | EXD1 | 70 | 41.294 | ENSPREG00000019468 | - | 72 | 40.796 | Poecilia_reticulata |
| ENSG00000178997 | EXD1 | 100 | 97.276 | ENSPPYG00000006369 | EXD1 | 100 | 97.276 | Pongo_abelii |
| ENSG00000178997 | EXD1 | 76 | 71.320 | ENSPCAG00000005269 | - | 76 | 71.574 | Procavia_capensis |
| ENSG00000178997 | EXD1 | 100 | 90.909 | ENSPCOG00000026598 | EXD1 | 100 | 82.398 | Propithecus_coquereli |
| ENSG00000178997 | EXD1 | 99 | 80.934 | ENSPVAG00000003149 | EXD1 | 99 | 80.934 | Pteropus_vampyrus |
| ENSG00000178997 | EXD1 | 68 | 43.038 | ENSPNAG00000008931 | exd1 | 76 | 43.038 | Pygocentrus_nattereri |
| ENSG00000178997 | EXD1 | 99 | 72.251 | ENSRNOG00000025360 | Exd1 | 99 | 72.251 | Rattus_norvegicus |
| ENSG00000178997 | EXD1 | 100 | 67.949 | ENSRBIG00000037257 | EXD1 | 100 | 96.875 | Rhinopithecus_bieti |
| ENSG00000178997 | EXD1 | 100 | 95.629 | ENSRROG00000034491 | EXD1 | 100 | 95.629 | Rhinopithecus_roxellana |
| ENSG00000178997 | EXD1 | 100 | 81.152 | ENSSBOG00000024842 | EXD1 | 100 | 81.152 | Saimiri_boliviensis_boliviensis |
| ENSG00000178997 | EXD1 | 92 | 63.956 | ENSSHAG00000016701 | EXD1 | 100 | 63.956 | Sarcophilus_harrisii |
| ENSG00000178997 | EXD1 | 66 | 44.000 | ENSSDUG00000004368 | exd1 | 63 | 43.501 | Seriola_dumerili |
| ENSG00000178997 | EXD1 | 99 | 75.146 | ENSSARG00000009556 | - | 99 | 75.146 | Sorex_araneus |
| ENSG00000178997 | EXD1 | 91 | 58.000 | ENSSPUG00000001716 | EXD1 | 73 | 52.048 | Sphenodon_punctatus |
| ENSG00000178997 | EXD1 | 100 | 90.909 | ENSSSCG00000004752 | EXD1 | 100 | 81.565 | Sus_scrofa |
| ENSG00000178997 | EXD1 | 67 | 38.677 | ENSTNIG00000017431 | - | 93 | 38.422 | Tetraodon_nigroviridis |
| ENSG00000178997 | EXD1 | 82 | 68.160 | ENSTBEG00000003085 | EXD1 | 99 | 68.160 | Tupaia_belangeri |
| ENSG00000178997 | EXD1 | 99 | 80.550 | ENSTTRG00000009809 | EXD1 | 99 | 80.550 | Tursiops_truncatus |
| ENSG00000178997 | EXD1 | 99 | 80.836 | ENSUMAG00000012170 | EXD1 | 99 | 80.836 | Ursus_maritimus |
| ENSG00000178997 | EXD1 | 99 | 80.350 | ENSVPAG00000007758 | EXD1 | 99 | 80.350 | Vicugna_pacos |
| ENSG00000178997 | EXD1 | 100 | 92.727 | ENSVVUG00000026704 | EXD1 | 100 | 79.826 | Vulpes_vulpes |
| ENSG00000178997 | EXD1 | 68 | 49.367 | ENSXETG00000017895 | exd1 | 95 | 48.861 | Xenopus_tropicalis |
| ENSG00000178997 | EXD1 | 58 | 42.598 | ENSXCOG00000010879 | - | 84 | 42.598 | Xiphophorus_couchianus |
| ENSG00000178997 | EXD1 | 61 | 42.816 | ENSXMAG00000014462 | - | 86 | 42.515 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | - | ISS | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0008408 | 3'-5' exonuclease activity | - | ISS | Function |
| GO:0031047 | gene silencing by RNA | - | ISS | Process |
| GO:0034587 | piRNA metabolic process | - | ISS | Process |
| GO:0042803 | protein homodimerization activity | - | ISS | Function |
| GO:0043186 | P granule | - | ISS | Component |
| GO:0051321 | meiotic cell cycle | - | IEA | Process |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | - | IEA | Process |
| GO:1990923 | PET complex | - | ISS | Component |