
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| DLBC | |||
| PCPG | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| BRCA | ||
| KIRP | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000554975 | SHMT2-216 | 780 | - | ENSP00000452404 | 215 (aa) | - | G3V5L0 |
| ENST00000553868 | SHMT2-208 | 941 | - | - | - (aa) | - | - |
| ENST00000557487 | SHMT2-232 | 1918 | - | ENSP00000452315 | 494 (aa) | - | P34897 |
| ENST00000556737 | SHMT2-224 | 572 | - | ENSP00000451495 | 147 (aa) | - | G3V3Y8 |
| ENST00000553950 | SHMT2-210 | 584 | - | - | - (aa) | - | - |
| ENST00000556689 | SHMT2-223 | 768 | - | ENSP00000452035 | 237 (aa) | - | G3V4W5 |
| ENST00000555213 | SHMT2-218 | 611 | - | - | - (aa) | - | - |
| ENST00000557302 | SHMT2-228 | 457 | - | - | - (aa) | - | - |
| ENST00000554656 | SHMT2-215 | 817 | - | - | - (aa) | - | - |
| ENST00000557740 | SHMT2-235 | 569 | - | - | - (aa) | - | - |
| ENST00000554467 | SHMT2-212 | 3416 | - | - | - (aa) | - | - |
| ENST00000557269 | SHMT2-227 | 576 | - | ENSP00000450594 | 52 (aa) | - | G3V2D2 |
| ENST00000557348 | SHMT2-229 | 1659 | - | ENSP00000450927 | 65 (aa) | - | G3V2Y1 |
| ENST00000449049 | SHMT2-203 | 2121 | - | ENSP00000413770 | 483 (aa) | - | P34897 |
| ENST00000555563 | SHMT2-219 | 895 | - | ENSP00000451630 | 86 (aa) | - | G3V3C6 |
| ENST00000554310 | SHMT2-211 | 579 | - | ENSP00000450893 | 160 (aa) | - | G3V2W0 |
| ENST00000556798 | SHMT2-225 | 525 | - | - | - (aa) | - | - |
| ENST00000553474 | SHMT2-205 | 1765 | - | ENSP00000452419 | 483 (aa) | - | P34897 |
| ENST00000557427 | SHMT2-230 | 569 | - | ENSP00000452045 | 159 (aa) | - | G3V4X0 |
| ENST00000553324 | SHMT2-204 | 566 | - | - | - (aa) | - | - |
| ENST00000557529 | SHMT2-233 | 1369 | - | ENSP00000450490 | 264 (aa) | - | H0YIZ0 |
| ENST00000328923 | SHMT2-201 | 2537 | - | ENSP00000333667 | 504 (aa) | - | P34897 |
| ENST00000554600 | SHMT2-213 | 837 | - | - | - (aa) | - | - |
| ENST00000555116 | SHMT2-217 | 1885 | - | ENSP00000452339 | 65 (aa) | - | G3V2Y1 |
| ENST00000554604 | SHMT2-214 | 879 | - | ENSP00000452163 | 192 (aa) | - | G3V540 |
| ENST00000556825 | SHMT2-226 | 2215 | - | ENSP00000451169 | 86 (aa) | - | G3V3C6 |
| ENST00000557703 | SHMT2-234 | 542 | - | ENSP00000450452 | 142 (aa) | - | G3V241 |
| ENST00000555634 | SHMT2-220 | 964 | - | ENSP00000450930 | 234 (aa) | - | G3V2Y4 |
| ENST00000553949 | SHMT2-209 | 552 | - | - | - (aa) | - | - |
| ENST00000557433 | SHMT2-231 | 1904 | - | ENSP00000450610 | 171 (aa) | - | G3V2E4 |
| ENST00000553837 | SHMT2-207 | 2152 | - | ENSP00000451371 | 192 (aa) | - | G3V540 |
| ENST00000553529 | SHMT2-206 | 562 | - | ENSP00000452161 | 147 (aa) | - | G3V3Y8 |
| ENST00000555774 | SHMT2-222 | 2311 | - | ENSP00000451872 | 65 (aa) | - | G3V2Y1 |
| ENST00000414700 | SHMT2-202 | 2006 | XM_011538675 | ENSP00000406881 | 483 (aa) | XP_011536977 | P34897 |
| ENST00000555773 | SHMT2-221 | 600 | - | ENSP00000451968 | 149 (aa) | - | G3V4T0 |
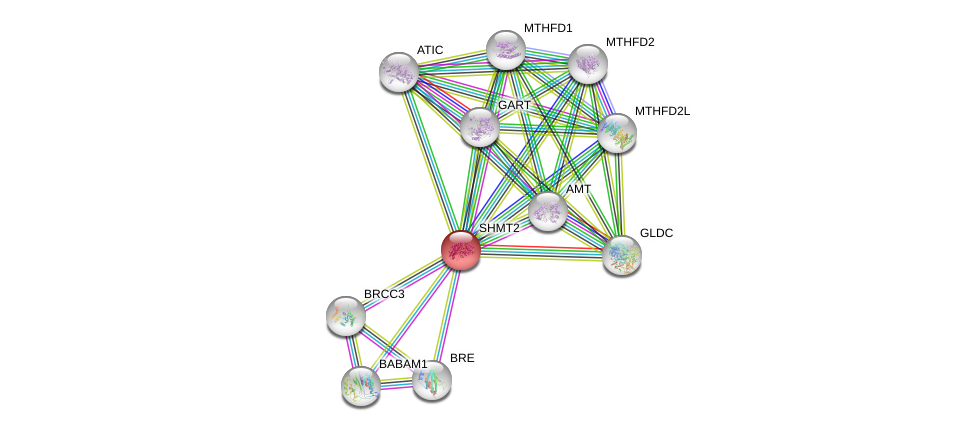
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000182199 | SHMT2 | 100 | 64.530 | FBgn0029823 | Shmt | 98 | 60.131 | Drosophila_melanogaster |
| ENSG00000182199 | SHMT2 | 100 | 95.814 | ENSMUSG00000025403 | Shmt2 | 99 | 94.036 | Mus_musculus |
| ENSG00000182199 | SHMT2 | 100 | 63.248 | YLR058C | SHM2 | 96 | 55.727 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002082 | regulation of oxidative phosphorylation | 21873635. | IBA | Process |
| GO:0002082 | regulation of oxidative phosphorylation | 29364879.29452640. | IMP | Process |
| GO:0003682 | chromatin binding | 18063578. | IDA | Function |
| GO:0004372 | glycine hydroxymethyltransferase activity | 21873635. | IBA | Function |
| GO:0004372 | glycine hydroxymethyltransferase activity | 8505317.17482557.24075985.25619277.29180469.29364879.29452640. | IDA | Function |
| GO:0005515 | protein binding | 25416956.25902260.26871637. | IPI | Function |
| GO:0005634 | nucleus | 24075985. | IDA | Component |
| GO:0005737 | cytoplasm | 24075985. | IDA | Component |
| GO:0005739 | mitochondrion | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | 17482557.24075985. | IDA | Component |
| GO:0005743 | mitochondrial inner membrane | 21873635. | IBA | Component |
| GO:0005743 | mitochondrial inner membrane | 21876188. | IDA | Component |
| GO:0005743 | mitochondrial inner membrane | - | TAS | Component |
| GO:0005758 | mitochondrial intermembrane space | - | IEA | Component |
| GO:0005759 | mitochondrial matrix | 21873635. | IBA | Component |
| GO:0005759 | mitochondrial matrix | 11516159.21876188. | IDA | Component |
| GO:0006544 | glycine metabolic process | 21873635. | IBA | Process |
| GO:0006544 | glycine metabolic process | 25619277.29180469. | IDA | Process |
| GO:0006563 | L-serine metabolic process | 25619277.29180469. | IDA | Process |
| GO:0006564 | L-serine biosynthetic process | - | IEA | Process |
| GO:0006565 | L-serine catabolic process | 21873635. | IBA | Process |
| GO:0006730 | one-carbon metabolic process | 21873635. | IBA | Process |
| GO:0006730 | one-carbon metabolic process | 11516159.29180469.29364879.29452640. | IDA | Process |
| GO:0008270 | zinc ion binding | 21873635. | IBA | Function |
| GO:0008284 | positive regulation of cell proliferation | - | IEA | Process |
| GO:0008732 | L-allo-threonine aldolase activity | - | IEA | Function |
| GO:0015630 | microtubule cytoskeleton | - | IDA | Component |
| GO:0016597 | amino acid binding | 21873635. | IBA | Function |
| GO:0019264 | glycine biosynthetic process from serine | 21873635. | IBA | Process |
| GO:0030170 | pyridoxal phosphate binding | 21873635. | IBA | Function |
| GO:0030170 | pyridoxal phosphate binding | 25619277. | IDA | Function |
| GO:0034340 | response to type I interferon | 21873635. | IBA | Process |
| GO:0034340 | response to type I interferon | 24075985. | IDA | Process |
| GO:0035999 | tetrahydrofolate interconversion | - | IEA | Process |
| GO:0042645 | mitochondrial nucleoid | 18063578. | IDA | Component |
| GO:0042802 | identical protein binding | 21873635. | IBA | Function |
| GO:0046653 | tetrahydrofolate metabolic process | 21873635. | IBA | Process |
| GO:0046653 | tetrahydrofolate metabolic process | 24075985.25619277. | IDA | Process |
| GO:0046655 | folic acid metabolic process | 21873635. | IBA | Process |
| GO:0046655 | folic acid metabolic process | - | TAS | Process |
| GO:0050897 | cobalt ion binding | 21873635. | IBA | Function |
| GO:0051262 | protein tetramerization | 25619277. | IDA | Process |
| GO:0051289 | protein homotetramerization | 21873635. | IBA | Process |
| GO:0051289 | protein homotetramerization | 29180469. | IDA | Process |
| GO:0070062 | extracellular exosome | 19056867.20458337. | HDA | Component |
| GO:0070129 | regulation of mitochondrial translation | 29364879.29452640. | IMP | Process |
| GO:0070536 | protein K63-linked deubiquitination | 24075985. | IMP | Process |
| GO:0070552 | BRISC complex | 21873635. | IBA | Component |
| GO:0070552 | BRISC complex | 24075985. | IDA | Component |
| GO:0070905 | serine binding | 21873635. | IBA | Function |
| GO:1903715 | regulation of aerobic respiration | 29364879.29452640. | IMP | Process |
| GO:1904482 | cellular response to tetrahydrofolate | 21873635. | IBA | Process |