
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 29417297 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| LGG | |||
| PAAD | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| MESO | ||
| KIRP | ||
| BLCA | ||
| LIHC | ||
| LGG | ||
| THCA | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000329918 | KH_1 | PF00013.29 | 2.1e-25 | 1 | 2 |
| ENSP00000329918 | KH_1 | PF00013.29 | 2.1e-25 | 2 | 2 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000329713 | MEX3B-201 | 3528 | - | ENSP00000329918 | 569 (aa) | - | Q6ZN04 |
| ENST00000558133 | MEX3B-202 | 4333 | - | ENSP00000456938 | 158 (aa) | - | Q6ZN04 |
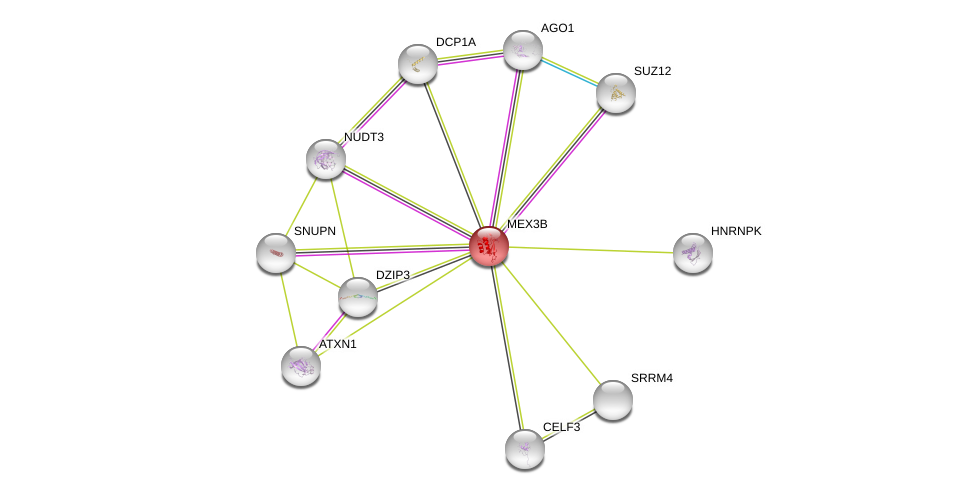
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000183496 | MEX3B | 90 | 59.216 | ENSG00000176624 | MEX3C | 89 | 59.216 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000183496 | MEX3B | 100 | 68.528 | ENSAPOG00000006457 | mex3b | 100 | 67.677 | Acanthochromis_polyacanthus |
| ENSG00000183496 | MEX3B | 96 | 90.128 | ENSAMEG00000006037 | MEX3B | 100 | 90.128 | Ailuropoda_melanoleuca |
| ENSG00000183496 | MEX3B | 59 | 83.251 | ENSACIG00000005655 | - | 64 | 84.772 | Amphilophus_citrinellus |
| ENSG00000183496 | MEX3B | 100 | 64.940 | ENSACIG00000017783 | mex3b | 100 | 63.385 | Amphilophus_citrinellus |
| ENSG00000183496 | MEX3B | 100 | 68.367 | ENSAOCG00000023903 | mex3b | 100 | 67.508 | Amphiprion_ocellaris |
| ENSG00000183496 | MEX3B | 100 | 68.367 | ENSAPEG00000000263 | mex3b | 100 | 67.508 | Amphiprion_percula |
| ENSG00000183496 | MEX3B | 100 | 68.213 | ENSATEG00000012352 | mex3b | 100 | 66.442 | Anabas_testudineus |
| ENSG00000183496 | MEX3B | 67 | 54.545 | ENSATEG00000015187 | - | 93 | 54.545 | Anabas_testudineus |
| ENSG00000183496 | MEX3B | 86 | 62.451 | ENSACAG00000011072 | MEX3B | 100 | 63.883 | Anolis_carolinensis |
| ENSG00000183496 | MEX3B | 100 | 97.902 | ENSANAG00000027164 | MEX3B | 100 | 97.902 | Aotus_nancymaae |
| ENSG00000183496 | MEX3B | 100 | 68.581 | ENSACLG00000025776 | mex3b | 100 | 67.622 | Astatotilapia_calliptera |
| ENSG00000183496 | MEX3B | 100 | 69.983 | ENSAMXG00000033615 | mex3b | 100 | 69.558 | Astyanax_mexicanus |
| ENSG00000183496 | MEX3B | 100 | 94.388 | ENSBTAG00000052904 | MEX3B | 100 | 94.388 | Bos_taurus |
| ENSG00000183496 | MEX3B | 100 | 94.889 | ENSCJAG00000008043 | MEX3B | 100 | 94.889 | Callithrix_jacchus |
| ENSG00000183496 | MEX3B | 100 | 97.364 | ENSCAFG00000013759 | MEX3B | 100 | 97.364 | Canis_familiaris |
| ENSG00000183496 | MEX3B | 100 | 95.525 | ENSCAFG00020015832 | MEX3B | 100 | 95.525 | Canis_lupus_dingo |
| ENSG00000183496 | MEX3B | 100 | 92.955 | ENSCHIG00000019863 | MEX3B | 100 | 93.471 | Capra_hircus |
| ENSG00000183496 | MEX3B | 86 | 98.965 | ENSTSYG00000025636 | MEX3B | 92 | 98.965 | Carlito_syrichta |
| ENSG00000183496 | MEX3B | 67 | 94.595 | ENSCAPG00000017918 | MEX3B | 84 | 94.595 | Cavia_aperea |
| ENSG00000183496 | MEX3B | 100 | 76.094 | ENSCPOG00000008241 | MEX3B | 100 | 75.926 | Cavia_porcellus |
| ENSG00000183496 | MEX3B | 100 | 93.823 | ENSCCAG00000001259 | MEX3B | 100 | 93.823 | Cebus_capucinus |
| ENSG00000183496 | MEX3B | 100 | 99.297 | ENSCATG00000030784 | MEX3B | 100 | 99.297 | Cercocebus_atys |
| ENSG00000183496 | MEX3B | 96 | 97.243 | ENSCLAG00000009451 | MEX3B | 100 | 97.243 | Chinchilla_lanigera |
| ENSG00000183496 | MEX3B | 100 | 99.123 | ENSCSAG00000015153 | MEX3B | 100 | 99.123 | Chlorocebus_sabaeus |
| ENSG00000183496 | MEX3B | 100 | 76.792 | ENSCPBG00000007809 | MEX3B | 100 | 77.474 | Chrysemys_picta_bellii |
| ENSG00000183496 | MEX3B | 97 | 98.195 | ENSCANG00000034524 | MEX3B | 99 | 95.423 | Colobus_angolensis_palliatus |
| ENSG00000183496 | MEX3B | 100 | 96.534 | ENSCGRG00001016198 | Mex3b | 100 | 96.534 | Cricetulus_griseus_chok1gshd |
| ENSG00000183496 | MEX3B | 86 | 97.521 | ENSCGRG00000018461 | Mex3b | 100 | 97.521 | Cricetulus_griseus_crigri |
| ENSG00000183496 | MEX3B | 100 | 63.051 | ENSCSEG00000005852 | mex3b | 95 | 62.203 | Cynoglossus_semilaevis |
| ENSG00000183496 | MEX3B | 100 | 71.331 | ENSDARG00000058369 | mex3b | 100 | 70.769 | Danio_rerio |
| ENSG00000183496 | MEX3B | 100 | 96.842 | ENSDNOG00000043628 | MEX3B | 100 | 96.842 | Dasypus_novemcinctus |
| ENSG00000183496 | MEX3B | 100 | 52.464 | ENSEBUG00000013015 | mex3b | 100 | 53.770 | Eptatretus_burgeri |
| ENSG00000183496 | MEX3B | 100 | 53.184 | ENSEBUG00000007080 | mex3b | 100 | 51.020 | Eptatretus_burgeri |
| ENSG00000183496 | MEX3B | 100 | 95.172 | ENSEASG00005007105 | MEX3B | 100 | 95.172 | Equus_asinus_asinus |
| ENSG00000183496 | MEX3B | 100 | 93.132 | ENSECAG00000013497 | MEX3B | 100 | 93.132 | Equus_caballus |
| ENSG00000183496 | MEX3B | 100 | 68.167 | ENSELUG00000002503 | mex3b | 100 | 68.500 | Esox_lucius |
| ENSG00000183496 | MEX3B | 100 | 95.675 | ENSFCAG00000037513 | MEX3B | 100 | 95.675 | Felis_catus |
| ENSG00000183496 | MEX3B | 76 | 86.207 | ENSFALG00000011102 | MEX3B | 100 | 69.271 | Ficedula_albicollis |
| ENSG00000183496 | MEX3B | 72 | 96.795 | ENSFDAG00000018452 | MEX3B | 100 | 96.795 | Fukomys_damarensis |
| ENSG00000183496 | MEX3B | 71 | 68.327 | ENSFHEG00000022740 | - | 68 | 67.143 | Fundulus_heteroclitus |
| ENSG00000183496 | MEX3B | 84 | 52.518 | ENSGMOG00000016534 | mex3b | 52 | 84.462 | Gadus_morhua |
| ENSG00000183496 | MEX3B | 90 | 75.095 | ENSGALG00000032984 | MEX3B | 92 | 77.820 | Gallus_gallus |
| ENSG00000183496 | MEX3B | 100 | 57.292 | ENSGACG00000012530 | mex3b | 100 | 56.272 | Gasterosteus_aculeatus |
| ENSG00000183496 | MEX3B | 100 | 77.281 | ENSGAGG00000022839 | MEX3B | 100 | 78.866 | Gopherus_agassizii |
| ENSG00000183496 | MEX3B | 99 | 98.765 | ENSGGOG00000000597 | MEX3B | 100 | 98.765 | Gorilla_gorilla |
| ENSG00000183496 | MEX3B | 100 | 68.750 | ENSHBUG00000022683 | mex3b | 100 | 67.791 | Haplochromis_burtoni |
| ENSG00000183496 | MEX3B | 100 | 94.590 | ENSHGLG00000012504 | MEX3B | 100 | 95.113 | Heterocephalus_glaber_female |
| ENSG00000183496 | MEX3B | 96 | 96.520 | ENSHGLG00100008208 | MEX3B | 100 | 96.520 | Heterocephalus_glaber_male |
| ENSG00000183496 | MEX3B | 66 | 60.399 | ENSHCOG00000010812 | - | 64 | 69.343 | Hippocampus_comes |
| ENSG00000183496 | MEX3B | 100 | 60.929 | ENSHCOG00000007290 | mex3b | 100 | 61.618 | Hippocampus_comes |
| ENSG00000183496 | MEX3B | 100 | 68.484 | ENSIPUG00000023356 | mex3b | 100 | 68.782 | Ictalurus_punctatus |
| ENSG00000183496 | MEX3B | 96 | 98.358 | ENSSTOG00000006738 | MEX3B | 90 | 98.358 | Ictidomys_tridecemlineatus |
| ENSG00000183496 | MEX3B | 100 | 95.855 | ENSJJAG00000015411 | Mex3b | 100 | 95.855 | Jaculus_jaculus |
| ENSG00000183496 | MEX3B | 90 | 63.602 | ENSLBEG00000015131 | mex3b | 95 | 63.218 | Labrus_bergylta |
| ENSG00000183496 | MEX3B | 100 | 74.957 | ENSLACG00000015887 | MEX3B | 100 | 75.000 | Latimeria_chalumnae |
| ENSG00000183496 | MEX3B | 100 | 72.589 | ENSLOCG00000012858 | mex3b | 100 | 72.420 | Lepisosteus_oculatus |
| ENSG00000183496 | MEX3B | 100 | 92.845 | ENSLAFG00000006027 | MEX3B | 98 | 97.000 | Loxodonta_africana |
| ENSG00000183496 | MEX3B | 100 | 98.599 | ENSMFAG00000037608 | MEX3B | 100 | 98.599 | Macaca_fascicularis |
| ENSG00000183496 | MEX3B | 100 | 96.907 | ENSMMUG00000018639 | MEX3B | 100 | 96.907 | Macaca_mulatta |
| ENSG00000183496 | MEX3B | 100 | 98.604 | ENSMNEG00000043420 | MEX3B | 100 | 98.604 | Macaca_nemestrina |
| ENSG00000183496 | MEX3B | 97 | 99.091 | ENSMLEG00000037832 | MEX3B | 100 | 99.091 | Mandrillus_leucophaeus |
| ENSG00000183496 | MEX3B | 100 | 68.942 | ENSMAMG00000011325 | mex3b | 100 | 68.421 | Mastacembelus_armatus |
| ENSG00000183496 | MEX3B | 100 | 68.750 | ENSMZEG00005003082 | mex3b | 100 | 67.791 | Maylandia_zebra |
| ENSG00000183496 | MEX3B | 100 | 96.187 | ENSMAUG00000001211 | Mex3b | 100 | 96.187 | Mesocricetus_auratus |
| ENSG00000183496 | MEX3B | 100 | 96.690 | ENSMICG00000043733 | MEX3B | 100 | 96.690 | Microcebus_murinus |
| ENSG00000183496 | MEX3B | 100 | 97.033 | ENSMOCG00000019726 | Mex3b | 100 | 97.033 | Microtus_ochrogaster |
| ENSG00000183496 | MEX3B | 56 | 70.036 | ENSMMOG00000000120 | - | 69 | 83.495 | Mola_mola |
| ENSG00000183496 | MEX3B | 100 | 63.095 | ENSMMOG00000015373 | mex3b | 98 | 60.647 | Mola_mola |
| ENSG00000183496 | MEX3B | 100 | 88.136 | ENSMODG00000019387 | MEX3B | 100 | 88.814 | Monodelphis_domestica |
| ENSG00000183496 | MEX3B | 100 | 67.116 | ENSMALG00000010929 | mex3b | 77 | 66.555 | Monopterus_albus |
| ENSG00000183496 | MEX3B | 100 | 96.367 | MGP_CAROLIEiJ_G0029937 | Mex3b | 100 | 96.367 | Mus_caroli |
| ENSG00000183496 | MEX3B | 100 | 96.360 | ENSMUSG00000057706 | Mex3b | 100 | 96.360 | Mus_musculus |
| ENSG00000183496 | MEX3B | 100 | 96.360 | MGP_PahariEiJ_G0013158 | Mex3b | 100 | 96.360 | Mus_pahari |
| ENSG00000183496 | MEX3B | 100 | 96.187 | MGP_SPRETEiJ_G0031039 | Mex3b | 100 | 96.187 | Mus_spretus |
| ENSG00000183496 | MEX3B | 100 | 97.364 | ENSMPUG00000015493 | MEX3B | 100 | 97.364 | Mustela_putorius_furo |
| ENSG00000183496 | MEX3B | 100 | 96.021 | ENSNGAG00000010102 | Mex3b | 100 | 96.021 | Nannospalax_galili |
| ENSG00000183496 | MEX3B | 100 | 68.750 | ENSNBRG00000000526 | mex3b | 98 | 67.791 | Neolamprologus_brichardi |
| ENSG00000183496 | MEX3B | 56 | 71.269 | ENSNBRG00000012485 | - | 65 | 84.772 | Neolamprologus_brichardi |
| ENSG00000183496 | MEX3B | 100 | 96.491 | ENSNLEG00000036285 | MEX3B | 100 | 96.491 | Nomascus_leucogenys |
| ENSG00000183496 | MEX3B | 86 | 89.796 | ENSMEUG00000002402 | MEX3B | 96 | 90.467 | Notamacropus_eugenii |
| ENSG00000183496 | MEX3B | 93 | 85.150 | ENSOPRG00000009604 | MEX3B | 100 | 85.150 | Ochotona_princeps |
| ENSG00000183496 | MEX3B | 100 | 94.618 | ENSODEG00000015600 | MEX3B | 100 | 94.444 | Octodon_degus |
| ENSG00000183496 | MEX3B | 100 | 68.581 | ENSONIG00000003391 | mex3b | 100 | 67.622 | Oreochromis_niloticus |
| ENSG00000183496 | MEX3B | 89 | 50.672 | ENSONIG00000015037 | - | 63 | 84.772 | Oreochromis_niloticus |
| ENSG00000183496 | MEX3B | 100 | 80.565 | ENSOCUG00000011603 | MEX3B | 100 | 81.229 | Oryctolagus_cuniculus |
| ENSG00000183496 | MEX3B | 62 | 77.232 | ENSOMEG00000023339 | - | 50 | 65.625 | Oryzias_melastigma |
| ENSG00000183496 | MEX3B | 100 | 95.855 | ENSOGAG00000029664 | MEX3B | 100 | 95.855 | Otolemur_garnettii |
| ENSG00000183496 | MEX3B | 74 | 91.463 | ENSOARG00000014515 | MEX3B | 92 | 92.793 | Ovis_aries |
| ENSG00000183496 | MEX3B | 98 | 99.461 | ENSPPAG00000020407 | MEX3B | 99 | 99.461 | Pan_paniscus |
| ENSG00000183496 | MEX3B | 100 | 99.126 | ENSPTRG00000007373 | MEX3B | 100 | 99.126 | Pan_troglodytes |
| ENSG00000183496 | MEX3B | 100 | 99.121 | ENSPANG00000015634 | MEX3B | 100 | 99.121 | Papio_anubis |
| ENSG00000183496 | MEX3B | 100 | 68.099 | ENSPKIG00000018385 | mex3b | 100 | 68.386 | Paramormyrops_kingsleyae |
| ENSG00000183496 | MEX3B | 100 | 64.957 | ENSPKIG00000019969 | - | 100 | 65.577 | Paramormyrops_kingsleyae |
| ENSG00000183496 | MEX3B | 100 | 72.899 | ENSPSIG00000007979 | MEX3B | 100 | 73.288 | Pelodiscus_sinensis |
| ENSG00000183496 | MEX3B | 100 | 68.439 | ENSPMGG00000015808 | mex3b | 100 | 68.439 | Periophthalmus_magnuspinnatus |
| ENSG00000183496 | MEX3B | 64 | 75.107 | ENSPMGG00000017559 | - | 64 | 84.422 | Periophthalmus_magnuspinnatus |
| ENSG00000183496 | MEX3B | 96 | 46.374 | ENSPMAG00000000125 | mex3b | 99 | 47.070 | Petromyzon_marinus |
| ENSG00000183496 | MEX3B | 55 | 69.742 | ENSPLAG00000002303 | - | 59 | 68.773 | Poecilia_latipinna |
| ENSG00000183496 | MEX3B | 55 | 70.111 | ENSPMEG00000006448 | - | 59 | 69.145 | Poecilia_mexicana |
| ENSG00000183496 | MEX3B | 73 | 96.643 | ENSPPYG00000006723 | - | 79 | 96.403 | Pongo_abelii |
| ENSG00000183496 | MEX3B | 86 | 97.930 | ENSPCAG00000014150 | MEX3B | 100 | 97.930 | Procavia_capensis |
| ENSG00000183496 | MEX3B | 100 | 96.528 | ENSPCOG00000019807 | MEX3B | 100 | 96.528 | Propithecus_coquereli |
| ENSG00000183496 | MEX3B | 100 | 97.544 | ENSPVAG00000000093 | MEX3B | 100 | 97.544 | Pteropus_vampyrus |
| ENSG00000183496 | MEX3B | 99 | 68.591 | ENSPNYG00000019384 | mex3b | 95 | 68.838 | Pundamilia_nyererei |
| ENSG00000183496 | MEX3B | 100 | 70.000 | ENSPNAG00000002109 | mex3b | 93 | 84.615 | Pygocentrus_nattereri |
| ENSG00000183496 | MEX3B | 100 | 96.014 | ENSRNOG00000025142 | Mex3b | 100 | 96.014 | Rattus_norvegicus |
| ENSG00000183496 | MEX3B | 100 | 98.418 | ENSRBIG00000033099 | MEX3B | 100 | 98.418 | Rhinopithecus_bieti |
| ENSG00000183496 | MEX3B | 100 | 98.418 | ENSRROG00000041868 | MEX3B | 100 | 98.418 | Rhinopithecus_roxellana |
| ENSG00000183496 | MEX3B | 89 | 98.611 | ENSSBOG00000031708 | MEX3B | 100 | 98.611 | Saimiri_boliviensis_boliviensis |
| ENSG00000183496 | MEX3B | 100 | 64.942 | ENSSFOG00015016667 | mex3b | 100 | 66.054 | Scleropages_formosus |
| ENSG00000183496 | MEX3B | 95 | 67.279 | ENSSFOG00015016308 | mex3b | 98 | 66.788 | Scleropages_formosus |
| ENSG00000183496 | MEX3B | 100 | 68.020 | ENSSMAG00000017078 | mex3b | 100 | 67.061 | Scophthalmus_maximus |
| ENSG00000183496 | MEX3B | 100 | 68.866 | ENSSDUG00000013254 | mex3b | 100 | 67.905 | Seriola_dumerili |
| ENSG00000183496 | MEX3B | 91 | 50.551 | ENSSDUG00000022255 | - | 52 | 82.381 | Seriola_dumerili |
| ENSG00000183496 | MEX3B | 100 | 68.866 | ENSSLDG00000014620 | mex3b | 100 | 68.243 | Seriola_lalandi_dorsalis |
| ENSG00000183496 | MEX3B | 86 | 74.327 | ENSSARG00000006007 | MEX3B | 100 | 74.534 | Sorex_araneus |
| ENSG00000183496 | MEX3B | 100 | 69.374 | ENSSPAG00000011854 | mex3b | 100 | 68.519 | Stegastes_partitus |
| ENSG00000183496 | MEX3B | 100 | 97.405 | ENSSSCG00000038491 | MEX3B | 100 | 97.405 | Sus_scrofa |
| ENSG00000183496 | MEX3B | 94 | 65.574 | ENSTRUG00000000367 | mex3b | 98 | 65.574 | Takifugu_rubripes |
| ENSG00000183496 | MEX3B | 100 | 65.636 | ENSTNIG00000003398 | mex3b | 100 | 62.585 | Tetraodon_nigroviridis |
| ENSG00000183496 | MEX3B | 100 | 93.467 | ENSUAMG00000019302 | MEX3B | 100 | 93.467 | Ursus_americanus |
| ENSG00000183496 | MEX3B | 92 | 95.985 | ENSVVUG00000004955 | MEX3B | 85 | 95.985 | Vulpes_vulpes |
| ENSG00000183496 | MEX3B | 100 | 71.577 | ENSXETG00000027601 | mex3b | 100 | 71.231 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000932 | P-body | - | IEA | Component |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0005509 | calcium ion binding | - | ISS | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006468 | protein phosphorylation | - | ISS | Process |
| GO:0046777 | protein autophosphorylation | - | ISS | Process |