
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| CHOL |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| SARC | ||
| COAD | ||
| READ | ||
| UCEC | ||
| LGG |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000566224 | USP7-214 | 503 | - | - | - (aa) | - | - |
| ENST00000567113 | USP7-216 | 448 | - | - | - (aa) | - | - |
| ENST00000565455 | USP7-210 | 3554 | - | ENSP00000456258 | 47 (aa) | - | H3BRI4 |
| ENST00000563043 | USP7-206 | 332 | - | ENSP00000459594 | 68 (aa) | - | I3L2D8 |
| ENST00000569448 | USP7-220 | 698 | - | - | - (aa) | - | - |
| ENST00000569230 | USP7-219 | 569 | - | ENSP00000457237 | 117 (aa) | - | H3BTM1 |
| ENST00000542333 | USP7-203 | 1966 | - | ENSP00000439272 | 509 (aa) | - | F5H2X1 |
| ENST00000566004 | USP7-212 | 559 | - | ENSP00000456150 | 130 (aa) | - | H3BRA2 |
| ENST00000570256 | USP7-221 | 591 | - | - | - (aa) | - | - |
| ENST00000564117 | USP7-209 | 479 | - | ENSP00000457815 | 109 (aa) | - | H3BUV0 |
| ENST00000565883 | USP7-211 | 453 | - | - | - (aa) | - | - |
| ENST00000562051 | USP7-204 | 567 | - | - | - (aa) | - | - |
| ENST00000567329 | USP7-217 | 567 | - | - | - (aa) | - | - |
| ENST00000344836 | USP7-201 | 5412 | XM_017023652 | ENSP00000343535 | 1102 (aa) | XP_016879141 | Q93009 |
| ENST00000563085 | USP7-207 | 3194 | - | ENSP00000454795 | 909 (aa) | - | H3BND8 |
| ENST00000566131 | USP7-213 | 569 | - | - | - (aa) | - | - |
| ENST00000566273 | USP7-215 | 577 | - | ENSP00000455719 | 159 (aa) | - | H3BQD1 |
| ENST00000567692 | USP7-218 | 313 | - | ENSP00000458027 | 17 (aa) | - | H3BVA7 |
| ENST00000381886 | USP7-202 | 3587 | XM_017023653 | ENSP00000371310 | 1086 (aa) | XP_016879142 | Q93009 |
| ENST00000562615 | USP7-205 | 304 | - | - | - (aa) | - | - |
| ENST00000563961 | USP7-208 | 4533 | - | ENSP00000454362 | 91 (aa) | - | H3BMF6 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000187555 | rs183725 | 16 | 8954178 | C | Platelet count | 27863252 | [0.017-0.035] unit increase | 0.02599643 | EFO_0004309 |
| ENSG00000187555 | rs7190551 | 16 | 8938158 | A | Platelet distribution width | 27863252 | [0.024-0.043] unit increase | 0.0334153 | EFO_0007984 |
| ENSG00000187555 | rs11551183 | 16 | 8894835 | G | Macrophage Migration Inhibitory Factor levels | 27989323 | [0.22-0.53] SD units decrease | 0.3715 | EFO_0008221 |
| ENSG00000187555 | rs141361914 | 16 | 8941610 | ? | Eosinophil counts | 30595370 | EFO_0004842 | ||
| ENSG00000187555 | rs2304467 | 16 | 8894920 | G | Chronotype | 30696823 | 1.0239831 | EFO_0008328 |
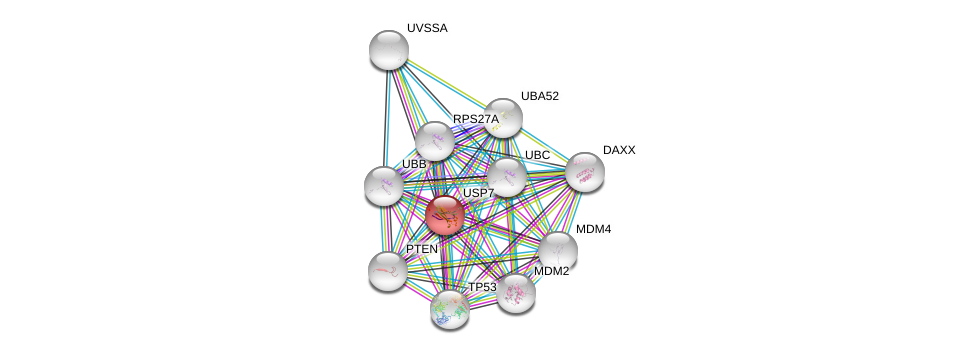
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000187555 | USP7 | 92 | 66.173 | FBgn0030366 | Usp7 | 95 | 48.584 | Drosophila_melanogaster |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002039 | p53 binding | 20096447. | IDA | Function |
| GO:0004197 | cysteine-type endopeptidase activity | 21873635. | IBA | Function |
| GO:0004197 | cysteine-type endopeptidase activity | 21745816. | IMP | Function |
| GO:0004843 | thiol-dependent ubiquitin-specific protease activity | 21873635. | IBA | Function |
| GO:0004843 | thiol-dependent ubiquitin-specific protease activity | 16964248.21745816.26280536. | IDA | Function |
| GO:0004843 | thiol-dependent ubiquitin-specific protease activity | 11923872. | IMP | Function |
| GO:0004843 | thiol-dependent ubiquitin-specific protease activity | - | TAS | Function |
| GO:0005515 | protein binding | 11279055.11923872.15916963.16402859.16474402.16713569.16845383.17268548.17936559.18566590.18716620.18729074.20601937.20697359.21170034.21745816.21988832.22056774.22466611.22466612.22653443.22810586.22863774.24823443.25172512.25266658.25544563.26496610.26678539.27123980. | IPI | Function |
| GO:0005634 | nucleus | 16791210. | HDA | Component |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 11279055.12093161.16964248.18410486.20096447.26678539. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005694 | chromosome | - | IEA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | 11279055. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006283 | transcription-coupled nucleotide-excision repair | 22466612. | IMP | Process |
| GO:0006283 | transcription-coupled nucleotide-excision repair | - | TAS | Process |
| GO:0006511 | ubiquitin-dependent protein catabolic process | - | IEA | Process |
| GO:0007275 | multicellular organism development | - | IEA | Process |
| GO:0008022 | protein C-terminus binding | 12093161. | IPI | Function |
| GO:0008134 | transcription factor binding | 21873635. | IBA | Function |
| GO:0008134 | transcription factor binding | 16964248. | IPI | Function |
| GO:0010216 | maintenance of DNA methylation | 21745816. | IMP | Process |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016567 | protein ubiquitination | - | TAS | Process |
| GO:0016579 | protein deubiquitination | 21873635. | IBA | Process |
| GO:0016579 | protein deubiquitination | 16964248.21745816.25172512. | IDA | Process |
| GO:0016579 | protein deubiquitination | 11923872.18410486.26678539.27123980. | IMP | Process |
| GO:0016579 | protein deubiquitination | - | TAS | Process |
| GO:0016604 | nuclear body | 16791210. | HDA | Component |
| GO:0016605 | PML body | - | IEA | Component |
| GO:0031625 | ubiquitin protein ligase binding | 11279055.18410486.18566590. | IPI | Function |
| GO:0031647 | regulation of protein stability | 21873635. | IBA | Process |
| GO:0031647 | regulation of protein stability | 27123980. | IDA | Process |
| GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 11279055. | IDA | Process |
| GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 26678539. | IMP | Process |
| GO:0032991 | protein-containing complex | 26678539. | IDA | Component |
| GO:0035520 | monoubiquitinated protein deubiquitination | 26280536. | IDA | Process |
| GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination | - | ISS | Process |
| GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity | 21745816. | IDA | Function |
| GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity | 26678539. | IMP | Function |
| GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity | - | TAS | Function |
| GO:0042752 | regulation of circadian rhythm | 27123980. | IDA | Process |
| GO:0050821 | protein stabilization | 25172512. | IDA | Process |
| GO:0050821 | protein stabilization | 18410486. | IMP | Process |
| GO:0051090 | regulation of DNA-binding transcription factor activity | 21873635. | IBA | Process |
| GO:0051090 | regulation of DNA-binding transcription factor activity | 16964248. | IDA | Process |
| GO:0071108 | protein K48-linked deubiquitination | - | IEA | Process |
| GO:0101005 | ubiquitinyl hydrolase activity | 18410486. | IMP | Function |
| GO:1901537 | positive regulation of DNA demethylation | 25944111. | IDA | Process |
| GO:1904353 | regulation of telomere capping | 25172512. | TAS | Process |
| GO:1990380 | Lys48-specific deubiquitinase activity | 25944111. | IDA | Function |