
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BRCA | |||
| ESCA | |||
| LAML | |||
| LGG | |||
| LIHC | |||
| SARC | |||
| STAD | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| LUSC | ||
| BRCA | ||
| ESCA | ||
| COAD | ||
| PAAD | ||
| BLCA | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| DLBC | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000343053 | NELFB-201 | 2698 | - | ENSP00000339495 | 628 (aa) | - | A0A0X1KG71 |
| ENST00000634710 | NELFB-202 | 1887 | - | ENSP00000489248 | 628 (aa) | - | Q8WX92 |
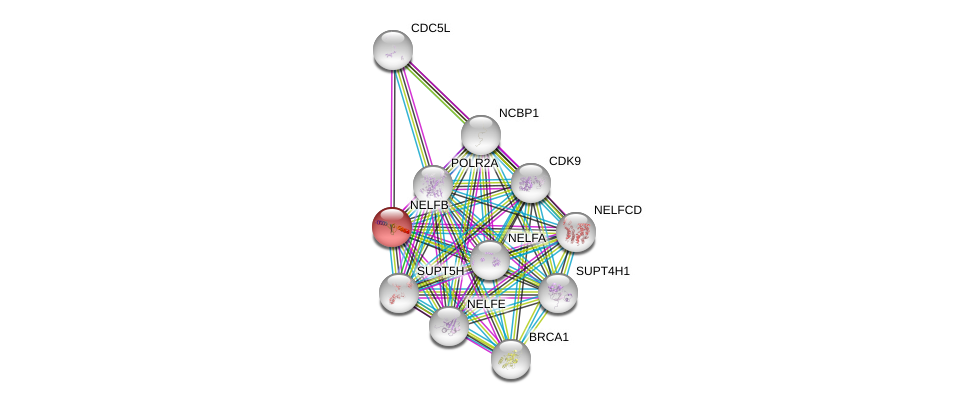
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | - | IEA | Function |
| GO:0005515 | protein binding | 11739404.12612062.14667819.17474147.20195357.26496610. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005737 | cytoplasm | - | IDA | Component |
| GO:0006366 | transcription by RNA polymerase II | - | TAS | Process |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | - | TAS | Process |
| GO:0008283 | cell proliferation | - | ISS | Process |
| GO:0032021 | NELF complex | 21873635. | IBA | Component |
| GO:0032021 | NELF complex | 12612062. | IDA | Component |
| GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter | 21873635. | IBA | Process |
| GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter | - | ISS | Process |
| GO:0048863 | stem cell differentiation | - | ISS | Process |
| GO:0050434 | positive regulation of viral transcription | - | TAS | Process |
| GO:2000737 | negative regulation of stem cell differentiation | - | ISS | Process |