
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PCPG | |||||||
| PRAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| PCPG | |||
| THCA | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| ACC | ||
| UCS | ||
| SKCM | ||
| PRAD | ||
| BRCA | ||
| ESCA | ||
| KIRP | ||
| PAAD | ||
| GBM | ||
| LGG | ||
| THCA | ||
| LUAD |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000340857 | H1F0-201 | 2344 | - | ENSP00000344504 | 194 (aa) | - | P07305 |
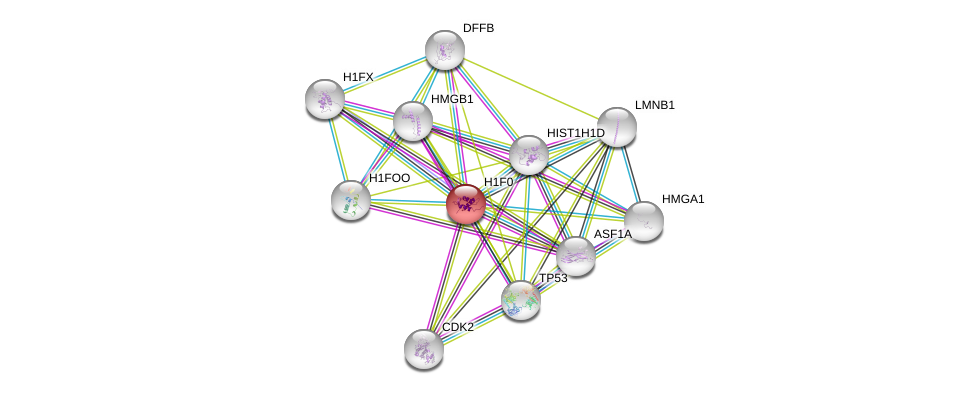
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000189060 | H1F0 | 100 | 94.845 | ENSMUSG00000096210 | H1f0 | 73 | 96.454 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 19158276. | IGI | Process |
| GO:0000786 | nucleosome | - | IEA | Component |
| GO:0000790 | nuclear chromatin | 21873635. | IBA | Component |
| GO:0000790 | nuclear chromatin | 19882353. | IDA | Component |
| GO:0003680 | AT DNA binding | - | IEA | Function |
| GO:0003690 | double-stranded DNA binding | 21873635. | IBA | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 9499053.26503038. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 21383955. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005719 | nuclear euchromatin | 21873635. | IBA | Component |
| GO:0005719 | nuclear euchromatin | 15911621. | IDA | Component |
| GO:0005794 | Golgi apparatus | - | IDA | Component |
| GO:0006309 | apoptotic DNA fragmentation | - | TAS | Process |
| GO:0006334 | nucleosome assembly | - | IEA | Process |
| GO:0006342 | chromatin silencing | 19158276. | IGI | Process |
| GO:0006355 | regulation of transcription, DNA-templated | 21873635. | IBA | Process |
| GO:0015629 | actin cytoskeleton | - | IDA | Component |
| GO:0016584 | nucleosome positioning | 21873635. | IBA | Process |
| GO:0016604 | nuclear body | 21873635. | IBA | Component |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0017053 | transcriptional repressor complex | 19158276. | IDA | Component |
| GO:0030261 | chromosome condensation | 21873635. | IBA | Process |
| GO:0031490 | chromatin DNA binding | 15911621. | IDA | Function |
| GO:0031492 | nucleosomal DNA binding | 21873635. | IBA | Function |
| GO:0031936 | negative regulation of chromatin silencing | 21873635. | IBA | Process |
| GO:0045910 | negative regulation of DNA recombination | 21873635. | IBA | Process |
| GO:2000679 | positive regulation of transcription regulatory region DNA binding | 19158276. | IMP | Process |