
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| KIRP | |||
| KIRP | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| SKCM | ||
| LUSC | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| BLCA | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000503346 | PDLIM7-209 | 669 | - | - | - (aa) | - | - |
| ENST00000355572 | PDLIM7-201 | 977 | - | ENSP00000347776 | 222 (aa) | - | Q9NR12 |
| ENST00000505074 | PDLIM7-213 | 931 | - | ENSP00000426213 | 287 (aa) | - | D6RH06 |
| ENST00000503827 | PDLIM7-210 | 451 | - | ENSP00000422363 | 120 (aa) | - | H0Y8W6 |
| ENST00000506537 | PDLIM7-216 | 636 | - | ENSP00000421664 | 90 (aa) | - | D6RAN1 |
| ENST00000486828 | PDLIM7-207 | 1904 | - | ENSP00000439157 | 191 (aa) | - | Q9NR12 |
| ENST00000504318 | PDLIM7-211 | 597 | - | - | - (aa) | - | - |
| ENST00000393551 | PDLIM7-205 | 1226 | - | ENSP00000377182 | 287 (aa) | - | Q9NR12 |
| ENST00000463411 | PDLIM7-206 | 1015 | - | - | - (aa) | - | - |
| ENST00000504380 | PDLIM7-212 | 556 | - | - | - (aa) | - | - |
| ENST00000393546 | PDLIM7-204 | 671 | - | ENSP00000377177 | 195 (aa) | - | H7BYK4 |
| ENST00000506161 | PDLIM7-215 | 759 | - | ENSP00000424850 | 173 (aa) | - | D6RF83 |
| ENST00000493815 | PDLIM7-208 | 1686 | - | ENSP00000431236 | 153 (aa) | - | Q9NR12 |
| ENST00000359895 | PDLIM7-203 | 1567 | - | ENSP00000352964 | 423 (aa) | - | Q9NR12 |
| ENST00000355841 | PDLIM7-202 | 1689 | XM_011534698 | ENSP00000348099 | 457 (aa) | XP_011533000 | Q9NR12 |
| ENST00000505746 | PDLIM7-214 | 405 | - | - | - (aa) | - | - |
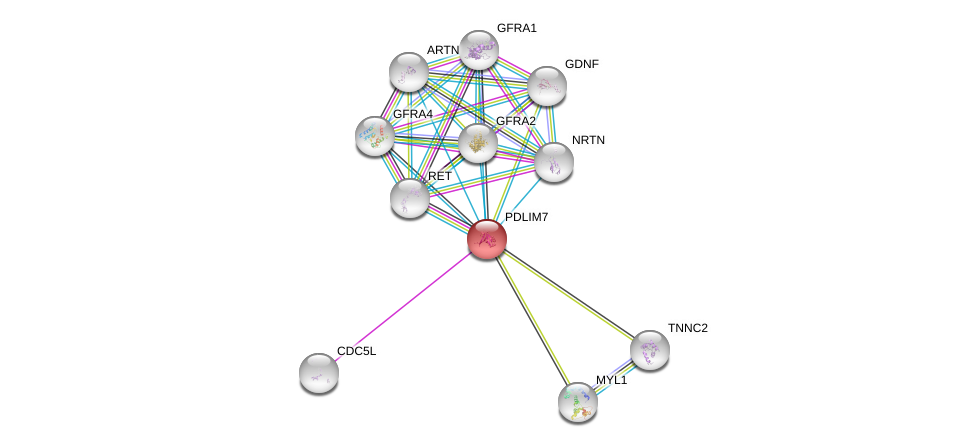
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001503 | ossification | - | IEA | Process |
| GO:0001725 | stress fiber | - | IEA | Component |
| GO:0001726 | ruffle | - | IEA | Component |
| GO:0005515 | protein binding | 16189514.16713569.23088713.25036637.25416956. | IPI | Function |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005913 | cell-cell adherens junction | 25468996. | IDA | Component |
| GO:0005925 | focal adhesion | - | IDA | Component |
| GO:0006898 | receptor-mediated endocytosis | 7929196. | TAS | Process |
| GO:0007411 | axon guidance | - | TAS | Process |
| GO:0015629 | actin cytoskeleton | - | IDA | Component |
| GO:0030036 | actin cytoskeleton organization | - | IEA | Process |
| GO:0045669 | positive regulation of osteoblast differentiation | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |