
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| KIRP | |||
| LAML | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| PRAD | ||
| LUSC | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| LGG | ||
| LUAD | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000522664 | ANXA6-213 | 451 | - | ENSP00000431086 | 91 (aa) | - | H0YC77 |
| ENST00000520054 | ANXA6-209 | 487 | - | - | - (aa) | - | - |
| ENST00000521749 | ANXA6-212 | 880 | - | ENSP00000430429 | 150 (aa) | - | E5RK63 |
| ENST00000521512 | ANXA6-211 | 1793 | - | ENSP00000430420 | 460 (aa) | - | E5RK69 |
| ENST00000523714 | ANXA6-215 | 2451 | - | ENSP00000430517 | 641 (aa) | - | P08133 |
| ENST00000520378 | ANXA6-210 | 1119 | - | - | - (aa) | - | - |
| ENST00000517677 | ANXA6-204 | 795 | - | ENSP00000430826 | 129 (aa) | - | E5RI05 |
| ENST00000523164 | ANXA6-214 | 923 | - | ENSP00000431078 | 110 (aa) | - | E5RJR0 |
| ENST00000517486 | ANXA6-203 | 562 | - | ENSP00000428916 | 156 (aa) | - | E5RFF0 |
| ENST00000517757 | ANXA6-206 | 553 | - | ENSP00000430572 | 129 (aa) | - | E5RJF5 |
| ENST00000354546 | ANXA6-201 | 2593 | XM_005268432 | ENSP00000346550 | 673 (aa) | - | P08133 |
| ENST00000377751 | ANXA6-202 | 1430 | - | ENSP00000366980 | 330 (aa) | - | E7EMC6 |
| ENST00000519644 | ANXA6-208 | 728 | - | ENSP00000430663 | 95 (aa) | - | E5RIU8 |
| ENST00000517707 | ANXA6-205 | 603 | - | - | - (aa) | - | - |
| ENST00000519610 | ANXA6-207 | 899 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000197043 | rs17728578 | 5 | 151138507 | ? | Adolescent idiopathic scoliosis | 30019117 | EFO_0005423 |
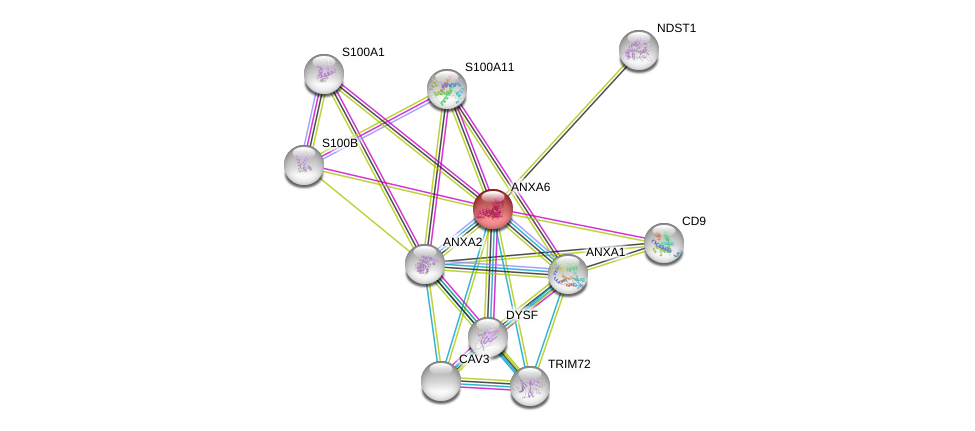
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000197043 | ANXA6 | 100 | 94.852 | ENSMUSG00000018340 | Anxa6 | 100 | 94.354 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001755 | neural crest cell migration | 21873635. | IBA | Process |
| GO:0001778 | plasma membrane repair | 21873635. | IBA | Process |
| GO:0001786 | phosphatidylserine binding | 21873635. | IBA | Function |
| GO:0003418 | growth plate cartilage chondrocyte differentiation | 21873635. | IBA | Process |
| GO:0005262 | calcium channel activity | 21873635. | IBA | Function |
| GO:0005509 | calcium ion binding | - | IEA | Function |
| GO:0005515 | protein binding | 15576473. | IPI | Function |
| GO:0005525 | GTP binding | 21873635. | IBA | Function |
| GO:0005525 | GTP binding | 16605264. | IMP | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 21873635. | IBA | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 2138016. | IDA | Function |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0005765 | lysosomal membrane | 23360953. | IDA | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0005925 | focal adhesion | 21873635. | IBA | Component |
| GO:0006937 | regulation of muscle contraction | 21873635. | IBA | Process |
| GO:0008201 | heparin binding | 21873635. | IBA | Function |
| GO:0008289 | lipid binding | 16605264. | IMP | Function |
| GO:0015276 | ligand-gated ion channel activity | 21873635. | IBA | Function |
| GO:0015276 | ligand-gated ion channel activity | 16605264. | IMP | Function |
| GO:0015485 | cholesterol binding | 21873635. | IBA | Function |
| GO:0015485 | cholesterol binding | 23360953. | IDA | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0019899 | enzyme binding | 21873635. | IBA | Function |
| GO:0031214 | biomineral tissue development | 21873635. | IBA | Process |
| GO:0031902 | late endosome membrane | 23360953. | IDA | Component |
| GO:0034220 | ion transmembrane transport | 16605264. | IMP | Process |
| GO:0035374 | chondroitin sulfate binding | 21873635. | IBA | Function |
| GO:0038023 | signaling receptor activity | 21873635. | IBA | Function |
| GO:0042470 | melanosome | - | IEA | Component |
| GO:0042803 | protein homodimerization activity | 21873635. | IBA | Function |
| GO:0042803 | protein homodimerization activity | 16605264. | IMP | Function |
| GO:0044877 | protein-containing complex binding | 21873635. | IBA | Function |
| GO:0048306 | calcium-dependent protein binding | 21873635. | IBA | Function |
| GO:0048306 | calcium-dependent protein binding | 15226301. | IPI | Function |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0051015 | actin filament binding | 21873635. | IBA | Function |
| GO:0051260 | protein homooligomerization | 21873635. | IBA | Process |
| GO:0051260 | protein homooligomerization | 16605264. | IMP | Process |
| GO:0051283 | negative regulation of sequestering of calcium ion | 21873635. | IBA | Process |
| GO:0051560 | mitochondrial calcium ion homeostasis | 21873635. | IBA | Process |
| GO:0062023 | collagen-containing extracellular matrix | 25037231.28344315.28675934. | HDA | Component |
| GO:0062023 | collagen-containing extracellular matrix | 28327460. | HDA | Component |
| GO:0070062 | extracellular exosome | 19056867.20458337.21362503.23533145. | HDA | Component |
| GO:0070509 | calcium ion import | 21873635. | IBA | Process |
| GO:0070588 | calcium ion transmembrane transport | - | IEA | Process |
| GO:0097190 | apoptotic signaling pathway | 21873635. | IBA | Process |