
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| OV | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| MESO | ||
| ACC | ||
| BRCA | ||
| PAAD | ||
| READ | ||
| DLBC | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000447319 | HMGN5-205 | 742 | - | ENSP00000408060 | 201 (aa) | - | Q5JSK9 |
| ENST00000373250 | HMGN5-202 | 572 | - | ENSP00000362347 | 161 (aa) | - | Q5JSK6 |
| ENST00000451455 | HMGN5-206 | 720 | - | ENSP00000404810 | 89 (aa) | - | Q5JSK7 |
| ENST00000430960 | HMGN5-203 | 578 | - | ENSP00000399626 | 148 (aa) | - | Q5JSL0 |
| ENST00000491275 | HMGN5-207 | 547 | - | - | - (aa) | - | - |
| ENST00000358130 | HMGN5-201 | 2126 | - | ENSP00000350848 | 282 (aa) | - | P82970 |
| ENST00000436386 | HMGN5-204 | 859 | - | ENSP00000413402 | 102 (aa) | - | Q5JSK8 |
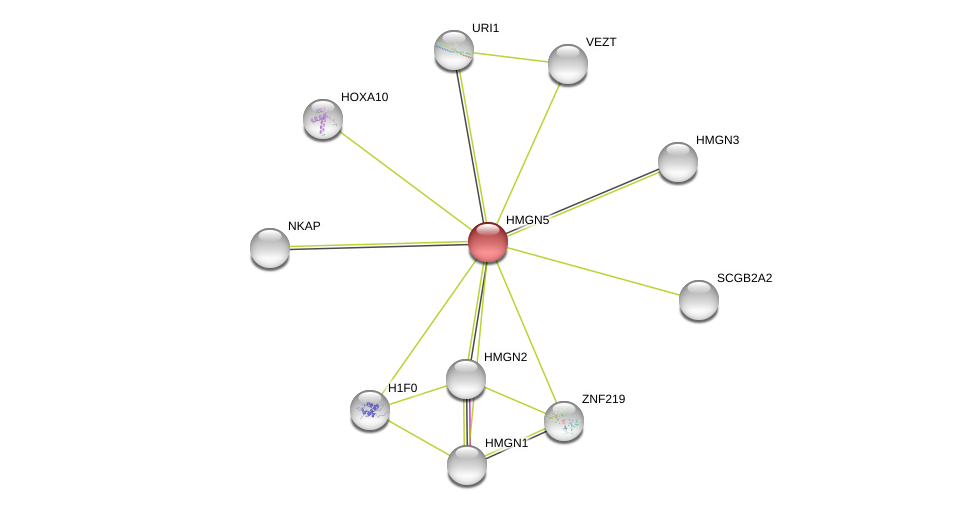
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000785 | chromatin | - | IEA | Component |
| GO:0003682 | chromatin binding | 11161810. | NAS | Function |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005634 | nucleus | 11161810. | NAS | Component |
| GO:0006325 | chromatin organization | - | IEA | Process |
| GO:0006355 | regulation of transcription, DNA-templated | 11161810. | NAS | Process |
| GO:0008284 | positive regulation of cell proliferation | 20531280. | IMP | Process |
| GO:0010628 | positive regulation of gene expression | 20531280. | IMP | Process |
| GO:0031492 | nucleosomal DNA binding | - | IEA | Function |
| GO:0043066 | negative regulation of apoptotic process | 20531280. | IMP | Process |
| GO:0045893 | positive regulation of transcription, DNA-templated | 11161810. | NAS | Process |
| GO:0071157 | negative regulation of cell cycle arrest | 20531280. | IMP | Process |