
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| KIRP | |||
| MESO | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| LUSC | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| BLCA | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LUAD | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000353793 | eIF2A | PF08662.11 | 2.6e-05 | 1 | 1 |
| ENSP00000399349 | eIF2A | PF08662.11 | 0.00014 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000567693 | WDHD1-205 | 2348 | - | ENSP00000456806 | 64 (aa) | - | H3BSQ1 |
| ENST00000475379 | WDHD1-204 | 696 | - | - | - (aa) | - | - |
| ENST00000420358 | WDHD1-202 | 3722 | XM_011536373 | ENSP00000399349 | 1006 (aa) | XP_011534675 | O75717 |
| ENST00000455555 | WDHD1-203 | 768 | - | ENSP00000413435 | 168 (aa) | - | C9JYB3 |
| ENST00000360586 | WDHD1-201 | 5996 | XM_006720012 | ENSP00000353793 | 1129 (aa) | XP_006720075 | O75717 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198554 | rs28489712 | 14 | 55017068 | ? | Response to anti-retroviral therapy (ddI/d4T) in HIV-1 infection (Grade 2 peripheral neuropathy) | 24554482 | [NR] | 31.7 | GO_0061479|EFO_0000180|EFO_0003100 |
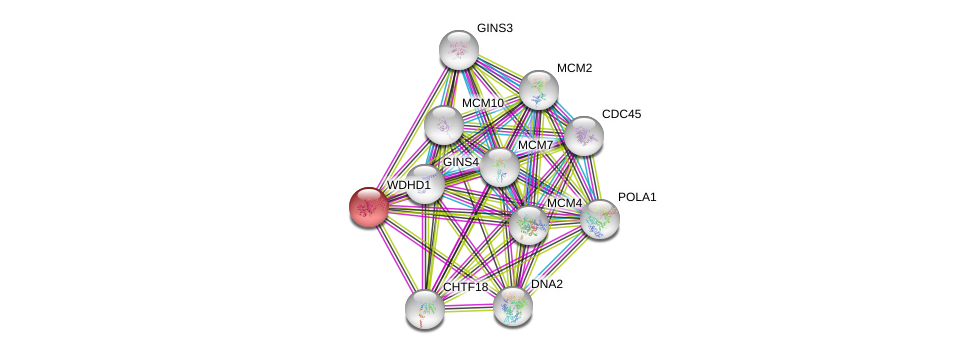
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSAPOG00000001560 | wdhd1 | 99 | 58.612 | Acanthochromis_polyacanthus |
| ENSG00000198554 | WDHD1 | 100 | 92.857 | ENSAMEG00000014356 | WDHD1 | 100 | 90.383 | Ailuropoda_melanoleuca |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSACIG00000021530 | wdhd1 | 99 | 58.547 | Amphilophus_citrinellus |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSAOCG00000018123 | wdhd1 | 99 | 58.983 | Amphiprion_ocellaris |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSAPEG00000000157 | wdhd1 | 99 | 59.103 | Amphiprion_percula |
| ENSG00000198554 | WDHD1 | 100 | 77.976 | ENSATEG00000021986 | wdhd1 | 99 | 59.075 | Anabas_testudineus |
| ENSG00000198554 | WDHD1 | 100 | 83.333 | ENSACAG00000016303 | WDHD1 | 99 | 66.551 | Anolis_carolinensis |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSACLG00000009036 | wdhd1 | 99 | 58.711 | Astatotilapia_calliptera |
| ENSG00000198554 | WDHD1 | 100 | 64.881 | ENSAMXG00000043712 | wdhd1 | 98 | 62.542 | Astyanax_mexicanus |
| ENSG00000198554 | WDHD1 | 100 | 95.238 | ENSBTAG00000019120 | WDHD1 | 100 | 88.771 | Bos_taurus |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSCJAG00000018208 | WDHD1 | 100 | 95.217 | Callithrix_jacchus |
| ENSG00000198554 | WDHD1 | 100 | 89.583 | ENSCAFG00000014989 | WDHD1 | 100 | 89.583 | Canis_familiaris |
| ENSG00000198554 | WDHD1 | 100 | 95.238 | ENSCHIG00000017956 | WDHD1 | 100 | 87.356 | Capra_hircus |
| ENSG00000198554 | WDHD1 | 100 | 96.429 | ENSTSYG00000000266 | WDHD1 | 100 | 89.213 | Carlito_syrichta |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSCCAG00000018389 | WDHD1 | 100 | 97.152 | Cebus_capucinus |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSCATG00000035221 | WDHD1 | 100 | 97.963 | Cercocebus_atys |
| ENSG00000198554 | WDHD1 | 100 | 92.262 | ENSCLAG00000013863 | - | 99 | 84.119 | Chinchilla_lanigera |
| ENSG00000198554 | WDHD1 | 100 | 97.321 | ENSCSAG00000014757 | WDHD1 | 100 | 97.288 | Chlorocebus_sabaeus |
| ENSG00000198554 | WDHD1 | 100 | 86.905 | ENSCPBG00000020071 | WDHD1 | 99 | 75.330 | Chrysemys_picta_bellii |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSCANG00000037721 | WDHD1 | 100 | 97.270 | Colobus_angolensis_palliatus |
| ENSG00000198554 | WDHD1 | 100 | 77.381 | ENSCVAG00000006106 | wdhd1 | 94 | 69.231 | Cyprinodon_variegatus |
| ENSG00000198554 | WDHD1 | 100 | 78.571 | ENSDARG00000015998 | wdhd1 | 97 | 69.153 | Danio_rerio |
| ENSG00000198554 | WDHD1 | 100 | 94.643 | ENSDNOG00000006661 | WDHD1 | 100 | 80.406 | Dasypus_novemcinctus |
| ENSG00000198554 | WDHD1 | 100 | 67.857 | ENSEASG00005003136 | - | 92 | 78.595 | Equus_asinus_asinus |
| ENSG00000198554 | WDHD1 | 100 | 92.857 | ENSECAG00000015412 | WDHD1 | 97 | 89.488 | Equus_caballus |
| ENSG00000198554 | WDHD1 | 100 | 77.976 | ENSELUG00000015575 | wdhd1 | 99 | 58.560 | Esox_lucius |
| ENSG00000198554 | WDHD1 | 100 | 92.857 | ENSFCAG00000023408 | - | 100 | 90.628 | Felis_catus |
| ENSG00000198554 | WDHD1 | 100 | 78.571 | ENSFHEG00000009177 | - | 96 | 70.764 | Fundulus_heteroclitus |
| ENSG00000198554 | WDHD1 | 100 | 85.119 | ENSGALG00000012199 | WDHD1 | 99 | 70.624 | Gallus_gallus |
| ENSG00000198554 | WDHD1 | 100 | 78.571 | ENSGACG00000017321 | wdhd1 | 99 | 49.520 | Gasterosteus_aculeatus |
| ENSG00000198554 | WDHD1 | 100 | 86.905 | ENSGAGG00000010665 | - | 99 | 79.147 | Gopherus_agassizii |
| ENSG00000198554 | WDHD1 | 100 | 99.405 | ENSGGOG00000007744 | WDHD1 | 100 | 99.203 | Gorilla_gorilla |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSHBUG00000015846 | - | 98 | 71.237 | Haplochromis_burtoni |
| ENSG00000198554 | WDHD1 | 100 | 91.071 | ENSHGLG00000015105 | WDHD1 | 100 | 84.262 | Heterocephalus_glaber_female |
| ENSG00000198554 | WDHD1 | 100 | 91.071 | ENSHGLG00100001967 | WDHD1 | 100 | 82.597 | Heterocephalus_glaber_male |
| ENSG00000198554 | WDHD1 | 100 | 77.976 | ENSHCOG00000007286 | wdhd1 | 100 | 55.958 | Hippocampus_comes |
| ENSG00000198554 | WDHD1 | 100 | 92.857 | ENSSTOG00000020300 | WDHD1 | 100 | 87.323 | Ictidomys_tridecemlineatus |
| ENSG00000198554 | WDHD1 | 100 | 77.723 | ENSJJAG00000001164 | Wdhd1 | 100 | 76.279 | Jaculus_jaculus |
| ENSG00000198554 | WDHD1 | 100 | 75.595 | ENSLBEG00000024847 | wdhd1 | 99 | 56.710 | Labrus_bergylta |
| ENSG00000198554 | WDHD1 | 99 | 75.000 | ENSLACG00000019053 | WDHD1 | 99 | 63.795 | Latimeria_chalumnae |
| ENSG00000198554 | WDHD1 | 100 | 82.143 | ENSLOCG00000012086 | wdhd1 | 99 | 59.478 | Lepisosteus_oculatus |
| ENSG00000198554 | WDHD1 | 100 | 94.048 | ENSLAFG00000005385 | WDHD1 | 100 | 88.654 | Loxodonta_africana |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSMFAG00000032915 | WDHD1 | 100 | 98.051 | Macaca_fascicularis |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSMMUG00000000302 | WDHD1 | 100 | 97.874 | Macaca_mulatta |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSMNEG00000032489 | WDHD1 | 100 | 98.051 | Macaca_nemestrina |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSMLEG00000035661 | WDHD1 | 100 | 98.051 | Mandrillus_leucophaeus |
| ENSG00000198554 | WDHD1 | 100 | 79.762 | ENSMAMG00000016742 | wdhd1 | 99 | 58.777 | Mastacembelus_armatus |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSMZEG00005015228 | wdhd1 | 99 | 58.885 | Maylandia_zebra |
| ENSG00000198554 | WDHD1 | 100 | 72.619 | ENSMZEG00005025682 | - | 99 | 65.079 | Maylandia_zebra |
| ENSG00000198554 | WDHD1 | 100 | 84.524 | ENSMGAG00000013676 | WDHD1 | 99 | 69.904 | Meleagris_gallopavo |
| ENSG00000198554 | WDHD1 | 100 | 95.833 | ENSMICG00000016239 | WDHD1 | 100 | 91.519 | Microcebus_murinus |
| ENSG00000198554 | WDHD1 | 100 | 77.976 | ENSMMOG00000007869 | wdhd1 | 99 | 55.429 | Mola_mola |
| ENSG00000198554 | WDHD1 | 100 | 85.119 | ENSMODG00000011703 | WDHD1 | 100 | 76.119 | Monodelphis_domestica |
| ENSG00000198554 | WDHD1 | 100 | 90.476 | MGP_CAROLIEiJ_G0019163 | Wdhd1 | 98 | 85.692 | Mus_caroli |
| ENSG00000198554 | WDHD1 | 100 | 89.881 | MGP_PahariEiJ_G0030180 | Wdhd1 | 98 | 85.220 | Mus_pahari |
| ENSG00000198554 | WDHD1 | 100 | 92.262 | ENSMPUG00000005542 | WDHD1 | 100 | 84.011 | Mustela_putorius_furo |
| ENSG00000198554 | WDHD1 | 100 | 93.452 | ENSMLUG00000010353 | WDHD1 | 100 | 88.771 | Myotis_lucifugus |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSNBRG00000010071 | wdhd1 | 99 | 59.123 | Neolamprologus_brichardi |
| ENSG00000198554 | WDHD1 | 100 | 99.405 | ENSNLEG00000012760 | WDHD1 | 100 | 94.066 | Nomascus_leucogenys |
| ENSG00000198554 | WDHD1 | 87 | 72.595 | ENSMEUG00000006706 | WDHD1 | 92 | 68.493 | Notamacropus_eugenii |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSONIG00000007084 | wdhd1 | 99 | 60.071 | Oreochromis_niloticus |
| ENSG00000198554 | WDHD1 | 100 | 88.690 | ENSOANG00000015129 | - | 100 | 83.521 | Ornithorhynchus_anatinus |
| ENSG00000198554 | WDHD1 | 100 | 95.238 | ENSOCUG00000004987 | WDHD1 | 100 | 88.614 | Oryctolagus_cuniculus |
| ENSG00000198554 | WDHD1 | 100 | 78.571 | ENSORLG00000019721 | wdhd1 | 99 | 56.734 | Oryzias_latipes |
| ENSG00000198554 | WDHD1 | 100 | 78.571 | ENSORLG00020007040 | wdhd1 | 99 | 57.756 | Oryzias_latipes_hni |
| ENSG00000198554 | WDHD1 | 100 | 78.571 | ENSORLG00015008848 | wdhd1 | 99 | 57.305 | Oryzias_latipes_hsok |
| ENSG00000198554 | WDHD1 | 100 | 91.071 | ENSOGAG00000003465 | WDHD1 | 100 | 84.704 | Otolemur_garnettii |
| ENSG00000198554 | WDHD1 | 100 | 95.238 | ENSOARG00000021081 | WDHD1 | 100 | 87.975 | Ovis_aries |
| ENSG00000198554 | WDHD1 | 100 | 99.405 | ENSPPAG00000035486 | WDHD1 | 100 | 96.457 | Pan_paniscus |
| ENSG00000198554 | WDHD1 | 100 | 92.857 | ENSPPRG00000007679 | WDHD1 | 100 | 90.539 | Panthera_pardus |
| ENSG00000198554 | WDHD1 | 100 | 92.262 | ENSPTIG00000013608 | WDHD1 | 100 | 90.363 | Panthera_tigris_altaica |
| ENSG00000198554 | WDHD1 | 100 | 99.702 | ENSPTRG00000006366 | WDHD1 | 100 | 99.646 | Pan_troglodytes |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSPANG00000017506 | WDHD1 | 100 | 97.874 | Papio_anubis |
| ENSG00000198554 | WDHD1 | 100 | 77.381 | ENSPKIG00000004863 | wdhd1 | 99 | 57.557 | Paramormyrops_kingsleyae |
| ENSG00000198554 | WDHD1 | 100 | 85.714 | ENSPSIG00000011635 | WDHD1 | 99 | 75.518 | Pelodiscus_sinensis |
| ENSG00000198554 | WDHD1 | 69 | 78.448 | ENSPLAG00000009199 | wdhd1 | 91 | 61.232 | Poecilia_latipinna |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSPPYG00000005834 | WDHD1 | 100 | 98.671 | Pongo_abelii |
| ENSG00000198554 | WDHD1 | 100 | 84.425 | ENSPVAG00000016064 | WDHD1 | 100 | 83.201 | Pteropus_vampyrus |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSPNYG00000019115 | wdhd1 | 94 | 70.903 | Pundamilia_nyererei |
| ENSG00000198554 | WDHD1 | 100 | 75.595 | ENSPNAG00000013823 | wdhd1 | 100 | 59.443 | Pygocentrus_nattereri |
| ENSG00000198554 | WDHD1 | 100 | 90.476 | ENSRNOG00000011167 | Wdhd1 | 100 | 81.416 | Rattus_norvegicus |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSRROG00000006993 | WDHD1 | 100 | 97.166 | Rhinopithecus_roxellana |
| ENSG00000198554 | WDHD1 | 100 | 98.810 | ENSSBOG00000034847 | WDHD1 | 100 | 90.523 | Saimiri_boliviensis_boliviensis |
| ENSG00000198554 | WDHD1 | 100 | 74.483 | ENSSHAG00000013551 | WDHD1 | 100 | 76.058 | Sarcophilus_harrisii |
| ENSG00000198554 | WDHD1 | 100 | 79.762 | ENSSFOG00015009950 | wdhd1 | 99 | 57.635 | Scleropages_formosus |
| ENSG00000198554 | WDHD1 | 100 | 77.976 | ENSSMAG00000007346 | wdhd1 | 99 | 56.707 | Scophthalmus_maximus |
| ENSG00000198554 | WDHD1 | 99 | 54.842 | ENSSDUG00000016996 | wdhd1 | 99 | 55.779 | Seriola_dumerili |
| ENSG00000198554 | WDHD1 | 100 | 77.976 | ENSSLDG00000021689 | wdhd1 | 98 | 67.722 | Seriola_lalandi_dorsalis |
| ENSG00000198554 | WDHD1 | 100 | 86.310 | ENSSPUG00000004721 | - | 94 | 80.537 | Sphenodon_punctatus |
| ENSG00000198554 | WDHD1 | 100 | 93.452 | ENSSSCG00000005052 | WDHD1 | 100 | 88.781 | Sus_scrofa |
| ENSG00000198554 | WDHD1 | 100 | 77.976 | ENSTRUG00000015390 | - | 97 | 69.281 | Takifugu_rubripes |
| ENSG00000198554 | WDHD1 | 100 | 75.595 | ENSTNIG00000018803 | wdhd1 | 99 | 57.407 | Tetraodon_nigroviridis |
| ENSG00000198554 | WDHD1 | 100 | 92.857 | ENSTTRG00000003314 | WDHD1 | 100 | 85.071 | Tursiops_truncatus |
| ENSG00000198554 | WDHD1 | 100 | 92.262 | ENSUAMG00000015360 | - | 96 | 91.715 | Ursus_americanus |
| ENSG00000198554 | WDHD1 | 100 | 92.262 | ENSUMAG00000015946 | WDHD1 | 100 | 89.567 | Ursus_maritimus |
| ENSG00000198554 | WDHD1 | 100 | 94.048 | ENSVPAG00000008394 | WDHD1 | 100 | 88.417 | Vicugna_pacos |
| ENSG00000198554 | WDHD1 | 100 | 90.476 | ENSVVUG00000029613 | WDHD1 | 100 | 88.230 | Vulpes_vulpes |
| ENSG00000198554 | WDHD1 | 100 | 79.167 | ENSXETG00000012210 | wdhd1 | 100 | 64.644 | Xenopus_tropicalis |
| ENSG00000198554 | WDHD1 | 100 | 77.381 | ENSXCOG00000000845 | - | 92 | 68.896 | Xiphophorus_couchianus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000278 | mitotic cell cycle | 21873635. | IBA | Process |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | ISM | Function |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0003682 | chromatin binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 21725360. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IEA | Component |
| GO:0005737 | cytoplasm | 9175701. | TAS | Component |
| GO:0006261 | DNA-dependent DNA replication | 21873635. | IBA | Process |
| GO:0006281 | DNA repair | 21873635. | IBA | Process |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 21873635. | IBA | Process |
| GO:0031298 | replication fork protection complex | 21873635. | IBA | Component |
| GO:0043596 | nuclear replication fork | 21873635. | IBA | Component |