
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| ACC | ||
| HNSC | ||
| PRAD | ||
| COAD | ||
| PCPG | ||
| BLCA | ||
| CESC | ||
| READ | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| THCA | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000410313 | ResIII | PF04851.15 | 1.4e-07 | 1 | 1 |
| ENSP00000392672 | ResIII | PF04851.15 | 1.5e-07 | 1 | 1 |
| ENSP00000399371 | ResIII | PF04851.15 | 2.2e-07 | 1 | 1 |
| ENSP00000416350 | ResIII | PF04851.15 | 2.9e-07 | 1 | 1 |
| ENSP00000379475 | ResIII | PF04851.15 | 4.8e-07 | 1 | 1 |
| ENSP00000416269 | ResIII | PF04851.15 | 4.8e-07 | 1 | 1 |
| ENSP00000365347 | ResIII | PF04851.15 | 5e-07 | 1 | 1 |
| ENSP00000392672 | DEAD | PF00270.29 | 7.6e-39 | 1 | 1 |
| ENSP00000399371 | DEAD | PF00270.29 | 1.3e-38 | 1 | 1 |
| ENSP00000410313 | DEAD | PF00270.29 | 1.3e-38 | 1 | 1 |
| ENSP00000379475 | DEAD | PF00270.29 | 2.5e-38 | 1 | 1 |
| ENSP00000416269 | DEAD | PF00270.29 | 2.5e-38 | 1 | 1 |
| ENSP00000365347 | DEAD | PF00270.29 | 2.8e-38 | 1 | 1 |
| ENSP00000416350 | DEAD | PF00270.29 | 9.8e-38 | 1 | 1 |
| ENSP00000408000 | DEAD | PF00270.29 | 6.9e-19 | 1 | 1 |
| ENSP00000409426 | DEAD | PF00270.29 | 5.1e-16 | 1 | 1 |
| ENSP00000405707 | DEAD | PF00270.29 | 2e-15 | 1 | 1 |
| ENSP00000393984 | DEAD | PF00270.29 | 3.2e-09 | 1 | 1 |
| ENSP00000399841 | DEAD | PF00270.29 | 1.6e-07 | 1 | 1 |
| ENSP00000405245 | DEAD | PF00270.29 | 2.9e-07 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000462256 | DDX39B-215 | 4027 | - | - | - (aa) | - | - |
| ENST00000481456 | DDX39B-219 | 5158 | - | - | - (aa) | - | - |
| ENST00000376177 | DDX39B-201 | 1529 | - | ENSP00000365347 | 425 (aa) | - | Q5STU3 |
| ENST00000474961 | DDX39B-217 | 974 | - | - | - (aa) | - | - |
| ENST00000484566 | DDX39B-221 | 857 | - | ENSP00000436220 | 141 (aa) | - | H0YCC6 |
| ENST00000456976 | DDX39B-213 | 720 | - | ENSP00000393984 | 125 (aa) | - | F6U6E2 |
| ENST00000458640 | DDX39B-214 | 2003 | - | ENSP00000416269 | 428 (aa) | - | Q13838 |
| ENST00000456662 | DDX39B-212 | 932 | - | ENSP00000416350 | 231 (aa) | - | F6S4E6 |
| ENST00000419338 | DDX39B-206 | 928 | - | ENSP00000410313 | 235 (aa) | - | A0A0A0MT12 |
| ENST00000428450 | DDX39B-209 | 633 | - | ENSP00000405707 | 197 (aa) | - | F6R6M7 |
| ENST00000417023 | DDX39B-203 | 561 | - | ENSP00000406422 | 187 (aa) | - | H0Y400 |
| ENST00000449757 | DDX39B-211 | 591 | - | ENSP00000409426 | 187 (aa) | - | F6QYI9 |
| ENST00000428098 | DDX39B-208 | 817 | - | ENSP00000392672 | 238 (aa) | - | F6TRA5 |
| ENST00000478365 | DDX39B-218 | 439 | - | - | - (aa) | - | - |
| ENST00000431908 | DDX39B-210 | 914 | - | ENSP00000408000 | 245 (aa) | - | F6UJC5 |
| ENST00000482195 | DDX39B-220 | 1232 | - | - | - (aa) | - | - |
| ENST00000419020 | DDX39B-205 | 583 | - | ENSP00000405245 | 132 (aa) | - | F6S2B7 |
| ENST00000462421 | DDX39B-216 | 618 | - | - | - (aa) | - | - |
| ENST00000396172 | DDX39B-202 | 2133 | - | ENSP00000379475 | 428 (aa) | - | Q13838 |
| ENST00000418897 | DDX39B-204 | 583 | - | ENSP00000399841 | 136 (aa) | - | F6SXL5 |
| ENST00000427214 | DDX39B-207 | 1005 | - | ENSP00000399371 | 289 (aa) | - | F6WLT2 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000198563 | Breast Neoplasms | 2.459E-13 | 17660530 |
| ENSG00000198563 | Diabetes Mellitus, Type 1 | 5.29E-68 | 19430480 |
| ENSG00000198563 | Behcet Syndrome | 4.55942540272892E-05 | 20622878 |
| ENSG00000198563 | Behcet Syndrome | 5.97381407474203E-06 | 20622878 |
| ENSG00000198563 | Behcet Syndrome | 8.82169288626833E-07 | 20622878 |
| ENSG00000198563 | Behcet Syndrome | 8.71917049005456E-07 | 20622878 |
| ENSG00000198563 | Behcet Syndrome | 1.03078253218308E-06 | 20622878 |
| ENSG00000198563 | Behcet Syndrome | 8.82169288626833E-07 | 20622878 |
| ENSG00000198563 | Behcet Syndrome | 1.0729589781576E-06 | 20622878 |
| ENSG00000198563 | Behcet Syndrome | 6.22624609680527E-07 | 20622878 |
| ENSG00000198563 | Platelet Function Tests | 8.1800000E-006 | - |
| ENSG00000198563 | Stevens-Johnson Syndrome | 2E-8 | 21912425 |
| ENSG00000198563 | Dermatitis, Atopic | 2E-6 | 22197932 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198563 | rs2269476 | 6 | 31540161 | A | Mean platelet volume | 27863252 | [0.039-0.057] unit increase | 0.04806173 | EFO_0004584 |
| ENSG00000198563 | rs114050967 | 6 | 31537703 | G | Myositis | 26291516 | [NR] | 2.32 | EFO_0000783 |
| ENSG00000198563 | rs9267485 | 6 | 31538013 | G | Mosquito bite size | 28199695 | [0.041-0.065] unit decrease | 0.0531442 | EFO_0008378 |
| ENSG00000198563 | rs34788230 | 6 | 31540676 | G | Monocyte percentage of white cells | 27863252 | [0.02-0.034] unit decrease | 0.02679379 | EFO_0007989 |
| ENSG00000198563 | rs2734583 | 6 | 31537703 | ? | Stevens-Johnson syndrome and toxic epidermal necrolysis (SJS-TEN) | 21912425 | [19.8-225.0] | 66.8 | EFO_0004276|EFO_0004775 |
| ENSG00000198563 | rs3853601 | 6 | 31531826 | G | Atopic dermatitis | 22197932 | [1.08-1.19] | 1.13 | EFO_0000274 |
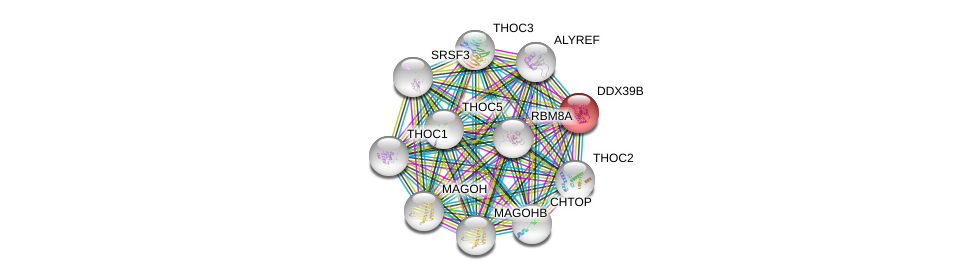
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000198563 | DDX39B | 78 | 30.256 | ENSG00000111364 | DDX55 | 85 | 31.902 |
| ENSG00000198563 | DDX39B | 100 | 89.953 | ENSG00000123136 | DDX39A | 100 | 89.953 |
| ENSG00000198563 | DDX39B | 91 | 32.832 | ENSG00000157349 | DDX19B | 98 | 33.243 |
| ENSG00000198563 | DDX39B | 80 | 38.095 | ENSG00000124228 | DDX27 | 88 | 31.837 |
| ENSG00000198563 | DDX39B | 86 | 32.367 | ENSG00000198231 | DDX42 | 51 | 30.685 |
| ENSG00000198563 | DDX39B | 81 | 32.821 | ENSG00000136271 | DDX56 | 88 | 35.976 |
| ENSG00000198563 | DDX39B | 96 | 38.547 | ENSG00000110367 | DDX6 | 79 | 35.659 |
| ENSG00000198563 | DDX39B | 83 | 31.120 | ENSG00000123064 | DDX54 | 91 | 32.812 |
| ENSG00000198563 | DDX39B | 95 | 31.351 | ENSG00000109832 | DDX25 | 94 | 40.625 |
| ENSG00000198563 | DDX39B | 84 | 32.922 | ENSG00000213782 | DDX47 | 75 | 35.159 |
| ENSG00000198563 | DDX39B | 91 | 36.257 | ENSG00000156976 | EIF4A2 | 79 | 50.000 |
| ENSG00000198563 | DDX39B | 91 | 36.842 | ENSG00000141543 | EIF4A3 | 88 | 38.356 |
| ENSG00000198563 | DDX39B | 93 | 30.387 | ENSG00000118197 | DDX59 | 79 | 30.387 |
| ENSG00000198563 | DDX39B | 80 | 30.612 | ENSG00000278053 | DDX52 | 50 | 41.379 |
| ENSG00000198563 | DDX39B | 92 | 37.687 | ENSG00000161960 | EIF4A1 | 95 | 41.406 |
| ENSG00000198563 | DDX39B | 87 | 36.053 | ENSG00000064703 | DDX20 | 51 | 39.437 |
| ENSG00000198563 | DDX39B | 91 | 32.581 | ENSG00000168872 | DDX19A | 95 | 33.246 |
| ENSG00000198563 | DDX39B | 87 | 31.759 | ENSG00000105671 | DDX49 | 71 | 39.241 |
| ENSG00000198563 | DDX39B | 77 | 30.029 | ENSG00000108654 | DDX5 | 70 | 30.081 |
| ENSG00000198563 | DDX39B | 99 | 30.116 | ENSG00000215301 | DDX3X | 79 | 30.052 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000198563 | DDX39B | 100 | 99.766 | ENSAMEG00000009508 | DDX39B | 99 | 99.766 | Ailuropoda_melanoleuca |
| ENSG00000198563 | DDX39B | 99 | 93.677 | ENSACIG00000004534 | ddx39b | 98 | 93.677 | Amphilophus_citrinellus |
| ENSG00000198563 | DDX39B | 100 | 92.824 | ENSATEG00000012366 | ddx39b | 100 | 92.824 | Anabas_testudineus |
| ENSG00000198563 | DDX39B | 100 | 97.203 | ENSACAG00000012240 | DDX39B | 100 | 97.203 | Anolis_carolinensis |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSANAG00000030372 | DDX39B | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000198563 | DDX39B | 95 | 95.506 | ENSACLG00000018759 | ddx39b | 99 | 90.783 | Astatotilapia_calliptera |
| ENSG00000198563 | DDX39B | 100 | 92.431 | ENSAMXG00000017837 | ddx39b | 100 | 92.431 | Astyanax_mexicanus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSBTAG00000014490 | DDX39B | 100 | 99.766 | Bos_taurus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCJAG00000003152 | DDX39B | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000198563 | DDX39B | 100 | 99.533 | ENSCAFG00000000509 | DDX39B | 100 | 100.000 | Canis_familiaris |
| ENSG00000198563 | DDX39B | 100 | 99.533 | ENSCAFG00020020530 | DDX39B | 100 | 99.533 | Canis_lupus_dingo |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCHIG00000004579 | DDX39B | 100 | 99.766 | Capra_hircus |
| ENSG00000198563 | DDX39B | 100 | 88.936 | ENSTSYG00000029741 | - | 91 | 88.095 | Carlito_syrichta |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSTSYG00000010226 | - | 100 | 100.000 | Carlito_syrichta |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCAPG00000010066 | DDX39B | 100 | 99.766 | Cavia_aperea |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCPOG00000002335 | DDX39B | 100 | 99.766 | Cavia_porcellus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCCAG00000027897 | DDX39B | 100 | 100.000 | Cebus_capucinus |
| ENSG00000198563 | DDX39B | 100 | 96.800 | ENSCATG00000043709 | DDX39B | 100 | 93.176 | Cercocebus_atys |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCLAG00000014555 | DDX39B | 100 | 99.766 | Chinchilla_lanigera |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCSAG00000008517 | DDX39B | 100 | 99.765 | Chlorocebus_sabaeus |
| ENSG00000198563 | DDX39B | 95 | 100.000 | ENSCPBG00000014671 | - | 100 | 100.000 | Chrysemys_picta_bellii |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSCANG00000041217 | DDX39B | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000198563 | DDX39B | 95 | 91.525 | ENSCGRG00001021916 | - | 89 | 84.358 | Cricetulus_griseus_chok1gshd |
| ENSG00000198563 | DDX39B | 100 | 99.160 | ENSCGRG00001017315 | - | 95 | 99.029 | Cricetulus_griseus_chok1gshd |
| ENSG00000198563 | DDX39B | 100 | 99.160 | ENSCGRG00000012479 | - | 76 | 99.184 | Cricetulus_griseus_crigri |
| ENSG00000198563 | DDX39B | 100 | 97.600 | ENSCGRG00000000078 | - | 90 | 98.537 | Cricetulus_griseus_crigri |
| ENSG00000198563 | DDX39B | 100 | 92.184 | ENSDARG00000036069 | ddx39b | 100 | 92.414 | Danio_rerio |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSDNOG00000010215 | DDX39B | 100 | 100.000 | Dasypus_novemcinctus |
| ENSG00000198563 | DDX39B | 100 | 99.654 | ENSDORG00000004985 | Ddx39b | 100 | 99.733 | Dipodomys_ordii |
| ENSG00000198563 | DDX39B | 100 | 98.701 | ENSETEG00000009649 | DDX39B | 87 | 86.898 | Echinops_telfairi |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSEASG00005017156 | DDX39B | 100 | 100.000 | Equus_asinus_asinus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSECAG00000007750 | DDX39B | 100 | 100.000 | Equus_caballus |
| ENSG00000198563 | DDX39B | 95 | 97.753 | ENSELUG00000017904 | ddx39b | 100 | 92.343 | Esox_lucius |
| ENSG00000198563 | DDX39B | 100 | 99.766 | ENSFCAG00000004497 | DDX39B | 100 | 99.766 | Felis_catus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSFDAG00000011074 | DDX39B | 100 | 99.766 | Fukomys_damarensis |
| ENSG00000198563 | DDX39B | 95 | 100.000 | ENSGAGG00000001300 | - | 80 | 99.684 | Gopherus_agassizii |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSGGOG00000005989 | DDX39B | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000198563 | DDX39B | 100 | 86.822 | ENSHBUG00000022054 | - | 77 | 88.038 | Haplochromis_burtoni |
| ENSG00000198563 | DDX39B | 100 | 99.299 | ENSHGLG00000019248 | DDX39B | 100 | 99.299 | Heterocephalus_glaber_female |
| ENSG00000198563 | DDX39B | 100 | 99.299 | ENSHGLG00100011850 | DDX39B | 100 | 99.299 | Heterocephalus_glaber_male |
| ENSG00000198563 | DDX39B | 95 | 96.629 | ENSIPUG00000015733 | DDX39B | 94 | 91.743 | Ictalurus_punctatus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSSTOG00000012838 | DDX39B | 100 | 99.533 | Ictidomys_tridecemlineatus |
| ENSG00000198563 | DDX39B | 100 | 99.299 | ENSJJAG00000005660 | - | 100 | 99.299 | Jaculus_jaculus |
| ENSG00000198563 | DDX39B | 100 | 76.050 | ENSLBEG00000017177 | - | 85 | 75.102 | Labrus_bergylta |
| ENSG00000198563 | DDX39B | 96 | 83.799 | ENSLBEG00000012254 | ddx39b | 99 | 78.220 | Labrus_bergylta |
| ENSG00000198563 | DDX39B | 95 | 100.000 | ENSLACG00000002110 | DDX39B | 100 | 89.862 | Latimeria_chalumnae |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSLAFG00000014202 | DDX39B | 100 | 99.766 | Loxodonta_africana |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSMFAG00000034233 | DDX39B | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSMMUG00000008839 | DDX39B | 100 | 100.000 | Macaca_mulatta |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSMNEG00000041059 | DDX39B | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSMLEG00000043791 | DDX39B | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000198563 | DDX39B | 95 | 95.506 | ENSMZEG00005017654 | ddx39b | 99 | 90.783 | Maylandia_zebra |
| ENSG00000198563 | DDX39B | 100 | 99.308 | ENSMAUG00000008059 | Ddx39b | 100 | 99.299 | Mesocricetus_auratus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSMICG00000017349 | DDX39B | 100 | 100.000 | Microcebus_murinus |
| ENSG00000198563 | DDX39B | 100 | 99.533 | ENSMOCG00000011169 | Ddx39b | 100 | 99.533 | Microtus_ochrogaster |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSMODG00000015791 | DDX39B | 100 | 95.294 | Monodelphis_domestica |
| ENSG00000198563 | DDX39B | 100 | 98.400 | MGP_CAROLIEiJ_G0021519 | - | 97 | 97.765 | Mus_caroli |
| ENSG00000198563 | DDX39B | 100 | 99.533 | ENSMUSG00000019432 | Ddx39b | 100 | 100.000 | Mus_musculus |
| ENSG00000198563 | DDX39B | 100 | 99.533 | MGP_PahariEiJ_G0020513 | Ddx39b | 100 | 100.000 | Mus_pahari |
| ENSG00000198563 | DDX39B | 100 | 99.533 | MGP_SPRETEiJ_G0022424 | Ddx39b | 100 | 99.533 | Mus_spretus |
| ENSG00000198563 | DDX39B | 100 | 99.766 | ENSMPUG00000011407 | DDX39B | 100 | 99.766 | Mustela_putorius_furo |
| ENSG00000198563 | DDX39B | 95 | 100.000 | ENSMLUG00000001681 | DDX39B | 100 | 100.000 | Myotis_lucifugus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSNGAG00000022948 | Ddx39b | 100 | 100.000 | Nannospalax_galili |
| ENSG00000198563 | DDX39B | 95 | 100.000 | ENSNLEG00000006094 | DDX39B | 100 | 75.467 | Nomascus_leucogenys |
| ENSG00000198563 | DDX39B | 100 | 98.400 | ENSMEUG00000008264 | DDX39B | 86 | 100.000 | Notamacropus_eugenii |
| ENSG00000198563 | DDX39B | 100 | 99.766 | ENSOPRG00000011923 | DDX39B | 100 | 99.766 | Ochotona_princeps |
| ENSG00000198563 | DDX39B | 100 | 99.567 | ENSODEG00000018083 | DDX39B | 100 | 99.533 | Octodon_degus |
| ENSG00000198563 | DDX39B | 100 | 91.898 | ENSONIG00000004740 | ddx39b | 100 | 91.898 | Oreochromis_niloticus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSOANG00000008361 | DDX39B | 95 | 99.156 | Ornithorhynchus_anatinus |
| ENSG00000198563 | DDX39B | 100 | 96.163 | ENSOCUG00000024126 | DDX39B | 100 | 96.163 | Oryctolagus_cuniculus |
| ENSG00000198563 | DDX39B | 95 | 100.000 | ENSOGAG00000009067 | - | 100 | 98.457 | Otolemur_garnettii |
| ENSG00000198563 | DDX39B | 100 | 98.621 | ENSOGAG00000028850 | - | 100 | 98.601 | Otolemur_garnettii |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSOARG00000008787 | DDX39B | 98 | 99.766 | Ovis_aries |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSPPAG00000043072 | DDX39B | 100 | 100.000 | Pan_paniscus |
| ENSG00000198563 | DDX39B | 100 | 99.766 | ENSPPRG00000001913 | DDX39B | 100 | 99.766 | Panthera_pardus |
| ENSG00000198563 | DDX39B | 100 | 98.832 | ENSPTIG00000017669 | DDX39B | 100 | 98.832 | Panthera_tigris_altaica |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSPTRG00000032509 | DDX39B | 100 | 100.000 | Pan_troglodytes |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSPANG00000018609 | DDX39B | 100 | 100.000 | Papio_anubis |
| ENSG00000198563 | DDX39B | 95 | 96.629 | ENSPKIG00000011324 | ddx39b | 100 | 91.304 | Paramormyrops_kingsleyae |
| ENSG00000198563 | DDX39B | 100 | 99.299 | ENSPEMG00000016905 | Ddx39b | 100 | 99.299 | Peromyscus_maniculatus_bairdii |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSPCIG00000017572 | DDX39B | 100 | 100.000 | Phascolarctos_cinereus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSPPYG00000016433 | DDX39B | 100 | 100.000 | Pongo_abelii |
| ENSG00000198563 | DDX39B | 95 | 99.438 | ENSPCAG00000003484 | DDX39B | 100 | 99.648 | Procavia_capensis |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSPCOG00000018656 | DDX39B | 100 | 100.000 | Propithecus_coquereli |
| ENSG00000198563 | DDX39B | 100 | 99.533 | ENSPVAG00000004653 | DDX39B | 100 | 99.533 | Pteropus_vampyrus |
| ENSG00000198563 | DDX39B | 94 | 95.429 | ENSPNYG00000010382 | ddx39b | 98 | 82.904 | Pundamilia_nyererei |
| ENSG00000198563 | DDX39B | 100 | 92.202 | ENSPNAG00000026926 | ddx39b | 100 | 92.202 | Pygocentrus_nattereri |
| ENSG00000198563 | DDX39B | 100 | 99.533 | ENSRNOG00000000841 | Ddx39b | 100 | 99.533 | Rattus_norvegicus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSRBIG00000031309 | DDX39B | 100 | 89.720 | Rhinopithecus_bieti |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSRROG00000039517 | DDX39B | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSSBOG00000035513 | - | 100 | 89.720 | Saimiri_boliviensis_boliviensis |
| ENSG00000198563 | DDX39B | 100 | 98.400 | ENSSBOG00000030486 | - | 97 | 97.945 | Saimiri_boliviensis_boliviensis |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSSHAG00000003209 | DDX39B | 100 | 100.000 | Sarcophilus_harrisii |
| ENSG00000198563 | DDX39B | 95 | 96.629 | ENSSFOG00015016107 | ddx39b | 100 | 91.762 | Scleropages_formosus |
| ENSG00000198563 | DDX39B | 100 | 93.287 | ENSSDUG00000011557 | ddx39b | 100 | 93.519 | Seriola_dumerili |
| ENSG00000198563 | DDX39B | 100 | 93.056 | ENSSLDG00000015763 | ddx39b | 100 | 93.287 | Seriola_lalandi_dorsalis |
| ENSG00000198563 | DDX39B | 100 | 99.160 | ENSSARG00000005245 | DDX39B | 100 | 87.701 | Sorex_araneus |
| ENSG00000198563 | DDX39B | 100 | 97.897 | ENSSPUG00000012805 | DDX39B | 100 | 97.897 | Sphenodon_punctatus |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSSSCG00000001400 | DDX39B | 100 | 100.000 | Sus_scrofa |
| ENSG00000198563 | DDX39B | 90 | 100.000 | ENSTBEG00000003721 | DDX39B | 100 | 76.869 | Tupaia_belangeri |
| ENSG00000198563 | DDX39B | 100 | 100.000 | ENSTTRG00000014074 | DDX39B | 100 | 91.455 | Tursiops_truncatus |
| ENSG00000198563 | DDX39B | 100 | 99.200 | ENSUAMG00000027335 | DDX39B | 100 | 86.977 | Ursus_americanus |
| ENSG00000198563 | DDX39B | 100 | 90.888 | ENSUMAG00000015124 | DDX39B | 100 | 90.888 | Ursus_maritimus |
| ENSG00000198563 | DDX39B | 90 | 100.000 | ENSVPAG00000009167 | - | 74 | 100.000 | Vicugna_pacos |
| ENSG00000198563 | DDX39B | 100 | 99.533 | ENSVVUG00000019518 | DDX39B | 99 | 99.533 | Vulpes_vulpes |
| ENSG00000198563 | DDX39B | 95 | 97.753 | ENSXETG00000002498 | ddx39b | 99 | 83.953 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000245 | spliceosomal complex assembly | 18593880. | IDA | Process |
| GO:0000346 | transcription export complex | 15833825.15998806. | IDA | Component |
| GO:0000398 | mRNA splicing, via spliceosome | 15047853. | IGI | Process |
| GO:0001889 | liver development | - | IEA | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0004004 | ATP-dependent RNA helicase activity | 17562711.18593880. | IDA | Function |
| GO:0005515 | protein binding | 11979277.14667819.15047853.15870275.19836239.21859714.23299939.30021884. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 21859714. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005681 | spliceosomal complex | 18593880. | IDA | Component |
| GO:0005687 | U4 snRNP | 18593880. | IDA | Component |
| GO:0005688 | U6 snRNP | 18593880. | IDA | Component |
| GO:0005737 | cytoplasm | 21859714. | IDA | Component |
| GO:0006405 | RNA export from nucleus | - | TAS | Process |
| GO:0006406 | mRNA export from nucleus | 17190602. | IDA | Process |
| GO:0006406 | mRNA export from nucleus | 15047853. | IGI | Process |
| GO:0008186 | RNA-dependent ATPase activity | 17562711. | IDA | Function |
| GO:0008380 | RNA splicing | 18593880. | IDA | Process |
| GO:0010501 | RNA secondary structure unwinding | 18593880. | IDA | Process |
| GO:0016363 | nuclear matrix | - | IEA | Component |
| GO:0016607 | nuclear speck | - | IDA | Component |
| GO:0016887 | ATPase activity | 20844015. | EXP | Function |
| GO:0017070 | U6 snRNA binding | 18593880. | IDA | Function |
| GO:0030621 | U4 snRNA binding | 18593880. | IDA | Function |
| GO:0031124 | mRNA 3'-end processing | - | TAS | Process |
| GO:0032786 | positive regulation of DNA-templated transcription, elongation | 22144908. | IMP | Process |
| GO:0042802 | identical protein binding | 14667819. | IPI | Function |
| GO:0043008 | ATP-dependent protein binding | 18593880. | IDA | Function |
| GO:0044877 | protein-containing complex binding | - | IEA | Function |
| GO:0045727 | positive regulation of translation | - | IEA | Process |
| GO:0046784 | viral mRNA export from host cell nucleus | 18974867. | IDA | Process |
| GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development | - | IEA | Process |
| GO:1904707 | positive regulation of vascular smooth muscle cell proliferation | - | IEA | Process |
| GO:2000002 | negative regulation of DNA damage checkpoint | 22144908. | IMP | Process |
| GO:2000573 | positive regulation of DNA biosynthetic process | - | IEA | Process |