
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| COAD | |||
| PAAD | |||
| READ | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| UCS | ||
| SKCM | ||
| LUSC | ||
| BRCA | ||
| COAD | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| CHOL | ||
| THCA | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000456849 | PAPSS2-202 | 3856 | - | ENSP00000406157 | 619 (aa) | - | O95340 |
| ENST00000482258 | PAPSS2-204 | 424 | - | - | - (aa) | - | - |
| ENST00000465996 | PAPSS2-203 | 275 | - | - | - (aa) | - | - |
| ENST00000361175 | PAPSS2-201 | 3949 | - | ENSP00000354436 | 614 (aa) | - | O95340 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000198682 | Exercise | 4E-6 | 19727025 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198682 | rs10887741 | 10 | 87683553 | T | Leisure-time exercise behaviour | 19727025 | [1.17-1.49] | 1.32 | EFO_0000483 |
| ENSG00000198682 | rs17173698 | 10 | 87709196 | ? | Heel bone mineral density | 30048462 | [0.044-0.07] unit decrease | 0.0574583 | EFO_0009270 |
| ENSG00000198682 | rs7896505 | 10 | 87717352 | ? | Heel bone mineral density | 30048462 | [0.013-0.021] unit increase | 0.0170634 | EFO_0009270 |
| ENSG00000198682 | rs17173698 | 10 | 87709196 | G | Heel bone mineral density | 30598549 | [0.028-0.051] unit decrease | 0.0391802 | EFO_0009270 |
| ENSG00000198682 | rs10887745 | 10 | 87722114 | G | Heel bone mineral density | 30598549 | [0.0097-0.0172] unit increase | 0.0134668 | EFO_0009270 |
| ENSG00000198682 | rs17173698 | 10 | 87709196 | ? | Heel bone mineral density | 30595370 | EFO_0009270 |
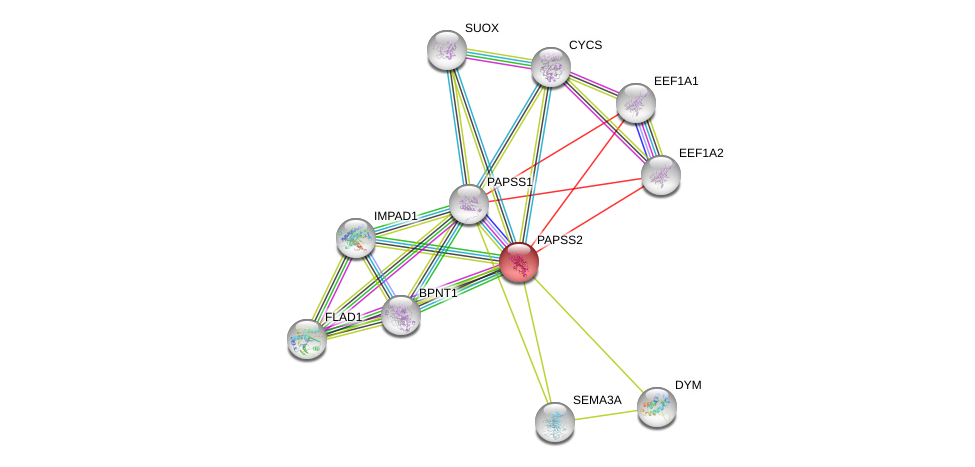
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000198682 | PAPSS2 | 98 | 78.369 | ENSG00000138801 | PAPSS1 | 96 | 78.369 | Homo_sapiens |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000103 | sulfate assimilation | 21873635. | IBA | Process |
| GO:0000103 | sulfate assimilation | - | IEA | Process |
| GO:0001501 | skeletal system development | 9771708. | TAS | Process |
| GO:0004020 | adenylylsulfate kinase activity | 21873635. | IBA | Function |
| GO:0004781 | sulfate adenylyltransferase (ATP) activity | - | ISS | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0007596 | blood coagulation | - | IEA | Process |
| GO:0016310 | phosphorylation | - | IEA | Process |
| GO:0016779 | nucleotidyltransferase activity | - | ISS | Function |
| GO:0050428 | 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process | 21873635. | IBA | Process |
| GO:0050428 | 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process | - | TAS | Process |
| GO:0060348 | bone development | - | IEA | Process |