
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BRCA | |||
| CESC | |||
| COAD | |||
| DLBC | |||
| LUAD | |||
| MESO | |||
| PRAD | |||
| READ | |||
| SKCM | |||
| STAD | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| BRCA | ||
| BLCA | ||
| UCEC | ||
| LIHC | ||
| UVM | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000359095 | MTS | PF05175.14 | 0.00041 | 1 | 1 |
| ENSP00000359095 | Met_10 | PF02475.16 | 0.00063 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000649727 | PRMT6-202 | 908 | - | - | - (aa) | - | - |
| ENST00000370078 | PRMT6-201 | 2616 | - | ENSP00000359095 | 375 (aa) | - | Q96LA8 |
| ENST00000650338 | PRMT6-203 | 2715 | - | ENSP00000497826 | 313 (aa) | - | - |
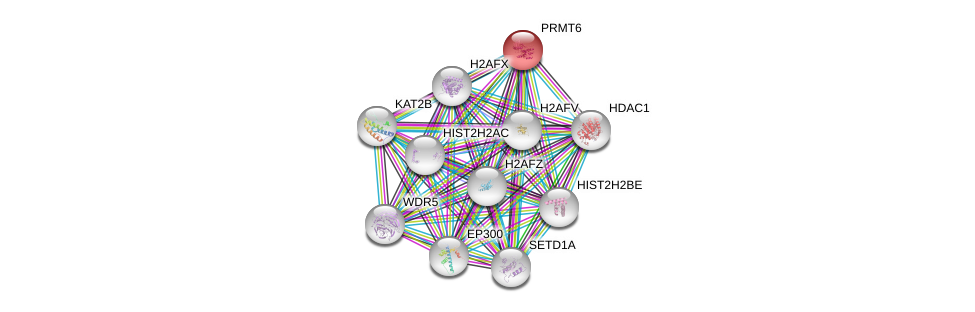
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000198890 | PRMT6 | 100 | 59.105 | ENSAPOG00000017409 | prmt6 | 95 | 58.017 | Acanthochromis_polyacanthus |
| ENSG00000198890 | PRMT6 | 100 | 55.096 | ENSACIG00000021067 | prmt6 | 94 | 55.689 | Amphilophus_citrinellus |
| ENSG00000198890 | PRMT6 | 100 | 57.508 | ENSAOCG00000023707 | prmt6 | 97 | 56.560 | Amphiprion_ocellaris |
| ENSG00000198890 | PRMT6 | 100 | 56.869 | ENSAPEG00000008762 | prmt6 | 97 | 55.977 | Amphiprion_percula |
| ENSG00000198890 | PRMT6 | 100 | 55.272 | ENSATEG00000021203 | prmt6 | 93 | 55.655 | Anabas_testudineus |
| ENSG00000198890 | PRMT6 | 94 | 94.898 | ENSANAG00000020517 | - | 100 | 88.000 | Aotus_nancymaae |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSANAG00000002736 | - | 100 | 99.733 | Aotus_nancymaae |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSACLG00000014684 | prmt6 | 93 | 56.456 | Astatotilapia_calliptera |
| ENSG00000198890 | PRMT6 | 100 | 59.105 | ENSAMXG00000025997 | prmt6 | 91 | 59.159 | Astyanax_mexicanus |
| ENSG00000198890 | PRMT6 | 100 | 93.610 | ENSBTAG00000039951 | PRMT6 | 100 | 92.533 | Bos_taurus |
| ENSG00000198890 | PRMT6 | 100 | 99.733 | ENSCJAG00000022625 | PRMT6 | 84 | 99.733 | Callithrix_jacchus |
| ENSG00000198890 | PRMT6 | 100 | 93.291 | ENSCHIG00000006399 | PRMT6 | 100 | 92.267 | Capra_hircus |
| ENSG00000198890 | PRMT6 | 100 | 96.805 | ENSCPOG00000030946 | PRMT6 | 100 | 94.933 | Cavia_porcellus |
| ENSG00000198890 | PRMT6 | 100 | 99.733 | ENSCCAG00000031625 | - | 100 | 99.733 | Cebus_capucinus |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSCATG00000043273 | PRMT6 | 100 | 99.467 | Cercocebus_atys |
| ENSG00000198890 | PRMT6 | 100 | 96.486 | ENSCLAG00000017949 | PRMT6 | 100 | 93.085 | Chinchilla_lanigera |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSCSAG00000000161 | PRMT6 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000198890 | PRMT6 | 100 | 96.486 | ENSCHOG00000007750 | PRMT6 | 100 | 94.933 | Choloepus_hoffmanni |
| ENSG00000198890 | PRMT6 | 99 | 46.984 | ENSCING00000001803 | - | 97 | 46.264 | Ciona_intestinalis |
| ENSG00000198890 | PRMT6 | 99 | 48.333 | ENSCSAVG00000002845 | - | 73 | 47.619 | Ciona_savignyi |
| ENSG00000198890 | PRMT6 | 100 | 99.681 | ENSCANG00000008600 | PRMT6 | 100 | 99.467 | Colobus_angolensis_palliatus |
| ENSG00000198890 | PRMT6 | 100 | 97.444 | ENSCGRG00001012887 | Prmt6 | 92 | 96.830 | Cricetulus_griseus_chok1gshd |
| ENSG00000198890 | PRMT6 | 100 | 58.147 | ENSCSEG00000013496 | prmt6 | 97 | 56.851 | Cynoglossus_semilaevis |
| ENSG00000198890 | PRMT6 | 100 | 55.272 | ENSCVAG00000002641 | prmt6 | 95 | 55.357 | Cyprinodon_variegatus |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSDARG00000069706 | prmt6 | 95 | 54.651 | Danio_rerio |
| ENSG00000198890 | PRMT6 | 100 | 94.888 | ENSDNOG00000002236 | PRMT6 | 100 | 93.600 | Dasypus_novemcinctus |
| ENSG00000198890 | PRMT6 | 100 | 92.332 | ENSDORG00000015482 | Prmt6 | 90 | 92.647 | Dipodomys_ordii |
| ENSG00000198890 | PRMT6 | 100 | 95.208 | ENSEASG00005022613 | PRMT6 | 100 | 94.400 | Equus_asinus_asinus |
| ENSG00000198890 | PRMT6 | 96 | 94.983 | ENSECAG00000003427 | PRMT6 | 95 | 92.265 | Equus_caballus |
| ENSG00000198890 | PRMT6 | 100 | 90.415 | ENSEEUG00000010272 | PRMT6 | 100 | 88.000 | Erinaceus_europaeus |
| ENSG00000198890 | PRMT6 | 100 | 57.188 | ENSELUG00000002245 | prmt6 | 94 | 56.725 | Esox_lucius |
| ENSG00000198890 | PRMT6 | 100 | 94.888 | ENSFCAG00000014050 | PRMT6 | 100 | 93.867 | Felis_catus |
| ENSG00000198890 | PRMT6 | 100 | 95.527 | ENSFDAG00000021017 | PRMT6 | 100 | 94.133 | Fukomys_damarensis |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSFHEG00000020586 | prmt6 | 95 | 56.250 | Fundulus_heteroclitus |
| ENSG00000198890 | PRMT6 | 100 | 52.866 | ENSGMOG00000020376 | prmt6 | 99 | 52.326 | Gadus_morhua |
| ENSG00000198890 | PRMT6 | 100 | 57.188 | ENSGAFG00000017234 | prmt6 | 95 | 57.143 | Gambusia_affinis |
| ENSG00000198890 | PRMT6 | 100 | 55.591 | ENSGACG00000002056 | prmt6 | 97 | 55.621 | Gasterosteus_aculeatus |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSGGOG00000028522 | PRMT6 | 100 | 99.733 | Gorilla_gorilla |
| ENSG00000198890 | PRMT6 | 100 | 55.591 | ENSHBUG00000015926 | prmt6 | 93 | 56.156 | Haplochromis_burtoni |
| ENSG00000198890 | PRMT6 | 100 | 97.125 | ENSHGLG00000009142 | PRMT6 | 100 | 94.933 | Heterocephalus_glaber_female |
| ENSG00000198890 | PRMT6 | 100 | 97.125 | ENSHGLG00100016337 | PRMT6 | 100 | 94.667 | Heterocephalus_glaber_male |
| ENSG00000198890 | PRMT6 | 100 | 53.185 | ENSHCOG00000017120 | prmt6 | 97 | 53.235 | Hippocampus_comes |
| ENSG00000198890 | PRMT6 | 100 | 57.188 | ENSIPUG00000002889 | prmt6 | 97 | 56.522 | Ictalurus_punctatus |
| ENSG00000198890 | PRMT6 | 100 | 97.764 | ENSSTOG00000010675 | PRMT6 | 100 | 96.533 | Ictidomys_tridecemlineatus |
| ENSG00000198890 | PRMT6 | 100 | 57.508 | ENSKMAG00000017402 | prmt6 | 93 | 57.440 | Kryptolebias_marmoratus |
| ENSG00000198890 | PRMT6 | 100 | 56.550 | ENSLOCG00000017743 | prmt6 | 94 | 57.057 | Lepisosteus_oculatus |
| ENSG00000198890 | PRMT6 | 100 | 97.444 | ENSLAFG00000025614 | PRMT6 | 100 | 96.533 | Loxodonta_africana |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSMFAG00000017156 | PRMT6 | 100 | 99.733 | Macaca_fascicularis |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSMMUG00000006802 | PRMT6 | 100 | 99.733 | Macaca_mulatta |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSMNEG00000037964 | PRMT6 | 100 | 99.733 | Macaca_nemestrina |
| ENSG00000198890 | PRMT6 | 100 | 99.681 | ENSMLEG00000009047 | PRMT6 | 100 | 99.467 | Mandrillus_leucophaeus |
| ENSG00000198890 | PRMT6 | 100 | 57.188 | ENSMAMG00000007319 | prmt6 | 98 | 56.232 | Mastacembelus_armatus |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSMZEG00005020230 | prmt6 | 93 | 56.456 | Maylandia_zebra |
| ENSG00000198890 | PRMT6 | 100 | 98.083 | ENSMICG00000016977 | PRMT6 | 100 | 96.543 | Microcebus_murinus |
| ENSG00000198890 | PRMT6 | 100 | 96.486 | ENSMOCG00000005923 | Prmt6 | 100 | 94.667 | Microtus_ochrogaster |
| ENSG00000198890 | PRMT6 | 100 | 53.994 | ENSMMOG00000005313 | prmt6 | 98 | 53.353 | Mola_mola |
| ENSG00000198890 | PRMT6 | 100 | 88.818 | ENSMODG00000002823 | PRMT6 | 88 | 89.189 | Monodelphis_domestica |
| ENSG00000198890 | PRMT6 | 100 | 93.291 | MGP_CAROLIEiJ_G0025550 | Prmt6 | 91 | 93.084 | Mus_caroli |
| ENSG00000198890 | PRMT6 | 100 | 94.249 | ENSMUSG00000049300 | Prmt6 | 92 | 93.660 | Mus_musculus |
| ENSG00000198890 | PRMT6 | 100 | 94.888 | MGP_PahariEiJ_G0026996 | Prmt6 | 92 | 94.236 | Mus_pahari |
| ENSG00000198890 | PRMT6 | 100 | 94.249 | MGP_SPRETEiJ_G0026500 | Prmt6 | 95 | 93.660 | Mus_spretus |
| ENSG00000198890 | PRMT6 | 100 | 94.569 | ENSMPUG00000019442 | PRMT6 | 100 | 93.600 | Mustela_putorius_furo |
| ENSG00000198890 | PRMT6 | 100 | 86.581 | ENSMLUG00000007086 | PRMT6 | 100 | 83.421 | Myotis_lucifugus |
| ENSG00000198890 | PRMT6 | 100 | 97.444 | ENSNGAG00000012858 | Prmt6 | 100 | 95.733 | Nannospalax_galili |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSNBRG00000012177 | prmt6 | 93 | 56.456 | Neolamprologus_brichardi |
| ENSG00000198890 | PRMT6 | 100 | 99.733 | ENSNLEG00000032446 | PRMT6 | 100 | 99.733 | Nomascus_leucogenys |
| ENSG00000198890 | PRMT6 | 100 | 89.137 | ENSMEUG00000000402 | PRMT6 | 92 | 87.755 | Notamacropus_eugenii |
| ENSG00000198890 | PRMT6 | 100 | 85.032 | ENSOPRG00000002067 | PRMT6 | 100 | 84.840 | Ochotona_princeps |
| ENSG00000198890 | PRMT6 | 100 | 94.249 | ENSODEG00000019666 | PRMT6 | 100 | 89.920 | Octodon_degus |
| ENSG00000198890 | PRMT6 | 97 | 55.410 | ENSONIG00000004768 | prmt6 | 91 | 55.657 | Oreochromis_niloticus |
| ENSG00000198890 | PRMT6 | 100 | 56.230 | ENSORLG00000005704 | prmt6 | 93 | 56.250 | Oryzias_latipes |
| ENSG00000198890 | PRMT6 | 100 | 56.230 | ENSORLG00020005446 | prmt6 | 93 | 56.250 | Oryzias_latipes_hni |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSORLG00015002130 | prmt6 | 95 | 55.952 | Oryzias_latipes_hsok |
| ENSG00000198890 | PRMT6 | 100 | 56.230 | ENSOMEG00000017676 | prmt6 | 95 | 56.250 | Oryzias_melastigma |
| ENSG00000198890 | PRMT6 | 100 | 96.805 | ENSOGAG00000033307 | PRMT6 | 100 | 96.000 | Otolemur_garnettii |
| ENSG00000198890 | PRMT6 | 100 | 93.291 | ENSOARG00000011289 | PRMT6 | 100 | 92.267 | Ovis_aries |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSPPAG00000013532 | PRMT6 | 100 | 100.000 | Pan_paniscus |
| ENSG00000198890 | PRMT6 | 100 | 95.208 | ENSPPRG00000021476 | PRMT6 | 100 | 94.133 | Panthera_pardus |
| ENSG00000198890 | PRMT6 | 100 | 94.888 | ENSPTIG00000005241 | PRMT6 | 100 | 93.867 | Panthera_tigris_altaica |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSPTRG00000001022 | PRMT6 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSPANG00000004113 | PRMT6 | 100 | 99.733 | Papio_anubis |
| ENSG00000198890 | PRMT6 | 100 | 60.383 | ENSPKIG00000008857 | prmt6 | 94 | 60.417 | Paramormyrops_kingsleyae |
| ENSG00000198890 | PRMT6 | 100 | 57.006 | ENSPMGG00000015586 | prmt6 | 94 | 56.105 | Periophthalmus_magnuspinnatus |
| ENSG00000198890 | PRMT6 | 100 | 97.444 | ENSPEMG00000014774 | Prmt6 | 100 | 95.200 | Peromyscus_maniculatus_bairdii |
| ENSG00000198890 | PRMT6 | 100 | 88.818 | ENSPCIG00000010181 | PRMT6 | 100 | 82.586 | Phascolarctos_cinereus |
| ENSG00000198890 | PRMT6 | 100 | 57.508 | ENSPFOG00000019926 | prmt6 | 95 | 57.738 | Poecilia_formosa |
| ENSG00000198890 | PRMT6 | 100 | 57.508 | ENSPLAG00000002189 | prmt6 | 93 | 57.738 | Poecilia_latipinna |
| ENSG00000198890 | PRMT6 | 100 | 57.508 | ENSPMEG00000017819 | prmt6 | 93 | 57.738 | Poecilia_mexicana |
| ENSG00000198890 | PRMT6 | 100 | 57.827 | ENSPREG00000010222 | prmt6 | 95 | 58.036 | Poecilia_reticulata |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSPPYG00000010685 | PRMT6 | 100 | 99.724 | Pongo_abelii |
| ENSG00000198890 | PRMT6 | 100 | 96.486 | ENSPCAG00000011732 | PRMT6 | 100 | 94.400 | Procavia_capensis |
| ENSG00000198890 | PRMT6 | 100 | 97.444 | ENSPCOG00000010650 | PRMT6 | 100 | 96.533 | Propithecus_coquereli |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSPNYG00000020634 | prmt6 | 93 | 56.456 | Pundamilia_nyererei |
| ENSG00000198890 | PRMT6 | 100 | 57.827 | ENSPNAG00000015218 | prmt6 | 86 | 56.322 | Pygocentrus_nattereri |
| ENSG00000198890 | PRMT6 | 100 | 95.847 | ENSRNOG00000017187 | Prmt6 | 100 | 93.867 | Rattus_norvegicus |
| ENSG00000198890 | PRMT6 | 100 | 95.847 | ENSRNOG00000048996 | LOC108348173 | 100 | 93.867 | Rattus_norvegicus |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSRBIG00000009563 | PRMT6 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000198890 | PRMT6 | 100 | 100.000 | ENSRROG00000021523 | PRMT6 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000198890 | PRMT6 | 100 | 98.722 | ENSSBOG00000018871 | PRMT6 | 100 | 98.667 | Saimiri_boliviensis_boliviensis |
| ENSG00000198890 | PRMT6 | 100 | 56.550 | ENSSMAG00000020345 | prmt6 | 96 | 54.810 | Scophthalmus_maximus |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSSDUG00000006628 | prmt6 | 96 | 54.942 | Seriola_dumerili |
| ENSG00000198890 | PRMT6 | 100 | 55.911 | ENSSLDG00000007963 | prmt6 | 95 | 55.102 | Seriola_lalandi_dorsalis |
| ENSG00000198890 | PRMT6 | 100 | 57.188 | ENSSPAG00000005623 | prmt6 | 97 | 56.268 | Stegastes_partitus |
| ENSG00000198890 | PRMT6 | 100 | 92.971 | ENSSSCG00000006853 | PRMT6 | 100 | 93.038 | Sus_scrofa |
| ENSG00000198890 | PRMT6 | 100 | 55.272 | ENSTRUG00000022052 | prmt6 | 96 | 55.389 | Takifugu_rubripes |
| ENSG00000198890 | PRMT6 | 100 | 55.272 | ENSTRUG00000019594 | prmt6 | 96 | 55.389 | Takifugu_rubripes |
| ENSG00000198890 | PRMT6 | 99 | 54.984 | ENSTNIG00000018964 | prmt6 | 99 | 53.959 | Tetraodon_nigroviridis |
| ENSG00000198890 | PRMT6 | 100 | 95.527 | ENSTBEG00000006318 | PRMT6 | 100 | 95.200 | Tupaia_belangeri |
| ENSG00000198890 | PRMT6 | 100 | 94.249 | ENSTTRG00000002369 | PRMT6 | 100 | 93.333 | Tursiops_truncatus |
| ENSG00000198890 | PRMT6 | 99 | 58.521 | ENSXETG00000009939 | prmt6 | 99 | 58.309 | Xenopus_tropicalis |
| ENSG00000198890 | PRMT6 | 100 | 56.869 | ENSXCOG00000005017 | prmt6 | 93 | 57.143 | Xiphophorus_couchianus |
| ENSG00000198890 | PRMT6 | 100 | 57.508 | ENSXMAG00000006494 | prmt6 | 95 | 57.738 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0003682 | chromatin binding | - | IEA | Function |
| GO:0005515 | protein binding | 16157300.16293633.16600869.23455924.25416956. | IPI | Function |
| GO:0005634 | nucleus | 11724789. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005730 | nucleolus | - | IDA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0006284 | base-excision repair | 16600869. | TAS | Process |
| GO:0006355 | regulation of transcription, DNA-templated | 21873635. | IBA | Process |
| GO:0008469 | histone-arginine N-methyltransferase activity | 19509293.22777353. | EXP | Function |
| GO:0008469 | histone-arginine N-methyltransferase activity | 21873635. | IBA | Function |
| GO:0008469 | histone-arginine N-methyltransferase activity | 23980157. | IDA | Function |
| GO:0008469 | histone-arginine N-methyltransferase activity | - | TAS | Function |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016274 | protein-arginine N-methyltransferase activity | - | TAS | Function |
| GO:0016571 | histone methylation | 19405910. | IDA | Process |
| GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine | 21873635. | IBA | Process |
| GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine | 17898714.18079182. | IDA | Process |
| GO:0034969 | histone arginine methylation | 21873635. | IBA | Process |
| GO:0034970 | histone H3-R2 methylation | 17898714.18079182. | IDA | Process |
| GO:0035241 | protein-arginine omega-N monomethyltransferase activity | 11724789.16157300.16600869. | IDA | Function |
| GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity | 21873635. | IBA | Function |
| GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity | 11724789.16157300.17898714.18079182. | IDA | Function |
| GO:0042054 | histone methyltransferase activity | 19405910. | IDA | Function |
| GO:0042393 | histone binding | 17898714.18079182. | IDA | Function |
| GO:0043985 | histone H4-R3 methylation | - | IEA | Process |
| GO:0044020 | histone methyltransferase activity (H4-R3 specific) | 18079182. | IDA | Function |
| GO:0045652 | regulation of megakaryocyte differentiation | - | TAS | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 18079182. | IDA | Process |
| GO:0051572 | negative regulation of histone H3-K4 methylation | - | IEA | Process |
| GO:0070611 | histone methyltransferase activity (H3-R2 specific) | 17898714.18079182. | IDA | Function |
| GO:0070612 | histone methyltransferase activity (H2A-R3 specific) | 18079182. | IDA | Function |
| GO:0090398 | cellular senescence | - | IEA | Process |
| GO:1901796 | regulation of signal transduction by p53 class mediator | - | IEA | Process |