| 9756848 | The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. | J Biol Chem | 1998 Oct 9 | Straub T | - |
| 9115577 | A frame shift mutation in a hot spot region of the nuclear autoantigen La (SS-B). | J Autoimmun | 1996 Dec | Bachmann M | - |
| 10574790 | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase. | Structure | 1999 Nov 15 | Li C | - |
| 30808650 | FUS (fused in sarcoma) is a component of the cellular response to topoisomerase I-induced DNA breakage and transcriptional stress. | Life Sci Alliance | 2019 Feb 26 | Martinez-Macias MI | doi: 10.26508/lsa.201800222 |
| 14585813 | Expression of nucleolar-related proteins in porcine preimplantation embryos produced in vivo and in vitro. | Biol Reprod | 2004 Apr | Bjerregaard B | - |
| 22207204 | The interface of transcription and DNA replication in the mitochondria. | Biochim Biophys Acta | 2012 Sep-Oct | Kasiviswanathan R | doi: 10.1016/j.bbagrm.2011.12.005 |
| 9588897 | Nucleolar function and size in cancer cells. | Am J Pathol | 1998 May | Derenzini M | - |
| 9211974 | Relative distribution of rDNA and proteins of the RNA polymerase I transcription machinery at chromosomal NORs. | Chromosoma | 1997 Jun | Suja JA | - |
| 12658636 | Nucleolar proteins and ultrastructure in bovine in vivo developed, in vitro produced, and parthenogenetic cleavage-stage embryos. | Mol Reprod Dev | 2003 May | Laurincik J | - |
| 11319160 | Immunolocalization of nucleolar proteins during bovine oocyte growth, meiotic maturation, and fertilization. | Biol Reprod | 2001 May | Fair T | - |
| 24667787 | Characterization of the transcriptional machinery bound across the widely presumed type 2 diabetes causal variant, rs7903146, within TCF7L2. | Eur J Hum Genet | 2015 Jan | Xia Q | doi: 10.1038/ejhg.2014.48 |
| 16427078 | Early effects of topoisomerase I inhibition on RNA polymerase II along transcribed genes in human cells. | J Mol Biol | 2006 Mar 17 | Khobta A | - |
| 6098156 | Enzyme studies of replication of the Escherichia coli chromosome. | Adv Exp Med Biol | 1984 | Kornberg A | - |
| 6088063 | Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. | Cell | 1984 Aug | Kaguni JM | - |
| 2175912 | Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. | Proc Natl Acad Sci U S A | 1990 Dec | Matsumoto T | - |
| 2181283 | Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. | Mol Cell Biol | 1990 Apr | Wang H | - |
| 9720989 | The phosphoprotein pp135 is an essential constituent of the fibrillar components of nucleoli and of coiled bodies. | Histochem Cell Biol | 1998 Aug | Vandelaer M | - |
| 9112231 | Antinuclear autoantibodies: probes for defining proteolytic events associated with apoptosis. | Mol Biol Rep | 1996 | Casiano CA | - |
| 8579976 | Immunologic aspects of scleroderma. | Curr Opin Rheumatol | 1995 Nov | White B | - |
| 8083304 | Conformational information in DNA: its role in the interaction with DNA topoisomerase I and nucleosomes. | J Cell Biochem | 1994 May | Caserta M | - |
| 11090457 | Nucleolar proteins and ultrastructure in preimplantation porcine embryos developed in vivo. | Biol Reprod | 2000 Dec | Hyttel P | - |
| 16226712 | Proteomic analysis of human O6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry. | Biochem Biophys Res Commun | 2005 Dec 2 | Niture SK | - |
| 15745627 | Meiosis and embryo technology: renaissance of the nucleolus. | Reprod Fertil Dev | 2005 | Maddox-Hyttel P | - |
| 15112326 | Nucleolar ultrastructure and protein allocation in in vitro produced porcine embryos. | Mol Reprod Dev | 2004 Jul | Laurincik J | - |
| 19132895 | Monoclonal antibody and siRNAs for topoisomerase I suppress telomerase activity. | Hybridoma (Larchmt) | 2009 Feb | Takahashi H | doi: 10.1089/hyb.2008.0066. |
| 18702506 | The binding of topoisomerase I to T antigen enhances the synthesis of RNA-DNA primers during simian virus 40 DNA replication. | Biochemistry | 2008 Sep 9 | Khopde S | doi: 10.1021/bi800825r |
| 17587596 | The giant fibrillar center: a nucleolar structure enriched in upstream binding factor (UBF) that appears in transcriptionally more active sensory ganglia neurons. | J Struct Biol | 2007 Sep | Casafont I | - |
| 17466364 | Ribosomal RNA and nucleolar proteins from the oocyte are to some degree used for embryonic nucleolar formation in cattle and pig. | Theriogenology | 2007 Sep 1 | Maddox-Hyttel P | - |
| 17176556 | Nucleolar remodeling in nuclear transfer embryos. | Adv Exp Med Biol | 2007 | Laurincik J | - |
| 21771901 | Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. | Proc Natl Acad Sci U S A | 2011 Aug 2 | Sperling AS | doi: 10.1073/pnas.1106834108 |
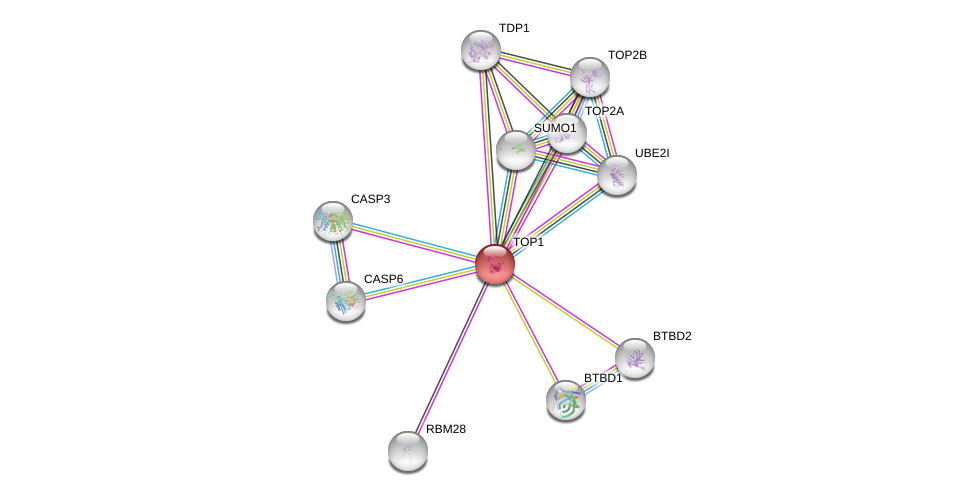
| 25851487 | RECQ5-dependent SUMOylation of DNA topoisomerase I prevents transcription-associated genome instability. | Nat Commun | 2015 Apr 8 | Li M | doi: 10.1038/ncomms7720. |