
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| ESCA | |||
| LGG | |||
| LIHC | |||
| PAAD | |||
| SKCM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| CESC | ||
| READ | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000394249 | PRC1-202 | 3034 | XM_006720759 | ENSP00000377793 | 620 (aa) | XP_006720822 | O43663 |
| ENST00000559811 | PRC1-215 | 414 | - | ENSP00000453117 | 110 (aa) | - | H0YLA0 |
| ENST00000559326 | PRC1-214 | 476 | - | - | - (aa) | - | - |
| ENST00000555455 | PRC1-206 | 830 | - | ENSP00000451344 | 238 (aa) | - | G3V3N7 |
| ENST00000417173 | PRC1-203 | 1215 | - | - | - (aa) | - | - |
| ENST00000559828 | PRC1-216 | 584 | - | ENSP00000453063 | 195 (aa) | - | H0YL53 |
| ENST00000361188 | PRC1-201 | 4087 | XM_005254987 | ENSP00000354679 | 606 (aa) | XP_005255044 | O43663 |
| ENST00000556129 | PRC1-209 | 583 | - | - | - (aa) | - | - |
| ENST00000557905 | PRC1-213 | 571 | - | ENSP00000453455 | 154 (aa) | - | H0YM42 |
| ENST00000555791 | PRC1-208 | 660 | - | - | - (aa) | - | - |
| ENST00000556972 | PRC1-210 | 728 | - | ENSP00000456737 | 46 (aa) | - | H3BSJ8 |
| ENST00000557763 | PRC1-212 | 933 | - | - | - (aa) | - | - |
| ENST00000560423 | PRC1-217 | 755 | - | ENSP00000453977 | 64 (aa) | - | H0YNE4 |
| ENST00000556982 | PRC1-211 | 583 | - | - | - (aa) | - | - |
| ENST00000560605 | PRC1-218 | 576 | - | - | - (aa) | - | - |
| ENST00000442656 | PRC1-204 | 2060 | XM_017022717 | ENSP00000409549 | 525 (aa) | XP_016878206 | O43663 |
| ENST00000560914 | PRC1-219 | 673 | - | - | - (aa) | - | - |
| ENST00000555745 | PRC1-207 | 582 | - | - | - (aa) | - | - |
| ENST00000553494 | PRC1-205 | 717 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198901 | rs12595025 | 15 | 90976901 | T | Mean platelet volume | 27863252 | [0.038-0.059] unit increase | 0.04882719 | EFO_0004584 |
| ENSG00000198901 | rs2290203 | 15 | 90968837 | G | Breast cancer | 25038754 | [1.05-1.11] | 1.08 | EFO_0000305 |
| ENSG00000198901 | rs8042680 | 15 | 90978107 | A | Type 2 diabetes | 20581827 | [1.05-1.09] | 1.07 | EFO_0001360 |
| ENSG00000198901 | rs8042680 | 15 | 90978107 | A | Type 2 diabetes | 28869590 | [0.039-0.075] unit increase | 0.057 | EFO_0001360 |
| ENSG00000198901 | rs8042680 | 15 | 90978107 | A | Type 2 diabetes | 28869590 | [0.042-0.084] unit increase | 0.0627 | EFO_0001360 |
| ENSG00000198901 | rs540970650 | 15 | 90973347 | ? | General cognitive ability | 29844566 | z-score decrease | 4.539 | EFO_0004337 |
| ENSG00000198901 | rs12910825 | 15 | 90968030 | G | Type 2 diabetes | 30054458 | [0.037-0.066] unit increase | 0.0517 | EFO_0001360 |
| ENSG00000198901 | rs2290203 | 15 | 90968837 | G | Breast cancer | 29059683 | [1.04-1.09] (Oncoarray) | 1.0638298 | EFO_0000305 |
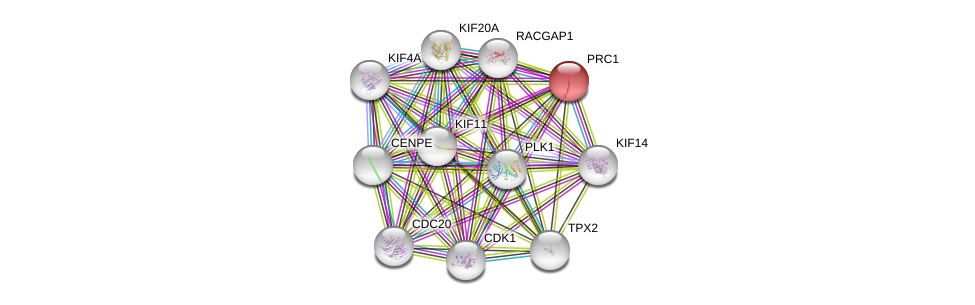
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000198901 | PRC1 | 100 | 84.416 | ENSMUSG00000038943 | Prc1 | 100 | 83.871 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000022 | mitotic spindle elongation | 9885575. | TAS | Process |
| GO:0000922 | spindle pole | - | IEA | Component |
| GO:0001578 | microtubule bundle formation | - | IEA | Process |
| GO:0005515 | protein binding | 14744859.16189514.16431929.17409436.19447967.19468300.23036704. | IPI | Function |
| GO:0005634 | nucleus | 17409436. | IDA | Component |
| GO:0005819 | spindle | 17351640. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005876 | spindle microtubule | 9885575. | TAS | Component |
| GO:0008017 | microtubule binding | - | IEA | Function |
| GO:0008284 | positive regulation of cell proliferation | 17409436. | IMP | Process |
| GO:0019894 | kinesin binding | 16431929. | IPI | Function |
| GO:0019901 | protein kinase binding | 17351640. | IPI | Function |
| GO:0030496 | midbody | 17409436. | IDA | Component |
| GO:0032465 | regulation of cytokinesis | 17351640. | IDA | Process |
| GO:0051301 | cell division | - | IEA | Process |
| GO:0070938 | contractile ring | 17409436. | IDA | Component |